













| General Information: |
|
| Name(s) found: |
AB36G_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1469 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrion [IDA] membrane [IDA] chloroplast envelope [IDA] plasma membrane [IDA] |
| Biological Process: |
defense response to fungus, incompatible interaction
[IMP]
callose deposition in cell wall during defense response [IMP] drug transmembrane transport [ISS] systemic acquired resistance [IMP] cadmium ion transport [IMP] indole glucosinolate catabolic process [IMP] negative regulation of defense response [IMP] defense response to bacterium [IMP] |
| Molecular Function: |
ATPase activity, coupled to transmembrane movement of substances
[ISS]
cadmium ion transmembrane transporter activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDYNPNLPPL GGGGVSMRRS ISRSVSRASR NIEDIFSSGS RRTQSVNDDE EALKWAAIEK 60
61 LPTYSRLRTT LMNAVVEDDV YGNQLMSKEV DVTKLDGEDR QKFIDMVFKV AEQDNERILT 120
121 KLRNRIDRVG IKLPTVEVRY EHLTIKADCY TGNRSLPTLL NVVRNMGESA LGMIGIQFAK 180
181 KAQLTILKDI SGVIKPGRMT LLLGPPSSGK TTLLLALAGK LDKSLQVSGD ITYNGYQLDE 240
241 FVPRKTSAYI SQNDLHVGIM TVKETLDFSA RCQGVGTRYD LLNELARREK DAGIFPEADV 300
301 DLFMKASAAQ GVKNSLVTDY TLKILGLDIC KDTIVGDDMM RGISGGQKKR VTTGEMIVGP 360
361 TKTLFMDEIS TGLDSSTTFQ IVKCLQQIVH LNEATVLMSL LQPAPETFDL FDDIILVSEG 420
421 QIVYQGPRDN ILEFFESFGF KCPERKGTAD FLQEVTSKKD QEQYWVNPNR PYHYIPVSEF 480
481 ASRYKSFHVG TKMSNELAVP FDKSRGHKAA LVFDKYSVSK RELLKSCWDK EWLLMQRNAF 540
541 FYVFKTVQIV IIAAITSTLF LRTEMNTRNE GDANLYIGAL LFGMIINMFN GFAEMAMMVS 600
601 RLPVFYKQRD LLFYPSWTFS LPTFLLGIPS SILESTAWMV VTYYSIGFAP DASRFFKQFL 660
661 LVFLIQQMAA SLFRLIASVC RTMMIANTGG ALTLLLVFLL GGFLLPKGKI PDWWGWAYWV 720
721 SPLTYAFNGL VVNEMFAPRW MNKMASSNST IKLGTMVLNT WDVYHQKNWY WISVGALLCF 780
781 TALFNILFTL ALTYLNPLGK KAGLLPEEEN EDADQGKDPM RRSLSTADGN RRGEVAMGRM 840
841 SRDSAAEASG GAGNKKGMVL PFTPLAMSFD DVKYFVDMPG EMRDQGVTET RLQLLKGVTG 900
901 AFRPGVLTAL MGVSGAGKTT LMDVLAGRKT GGYIEGDVRI SGFPKVQETF ARISGYCEQT 960
961 DIHSPQVTVR ESLIFSAFLR LPKEVGKDEK MMFVDQVMEL VELDSLRDSI VGLPGVTGLS1020
1021 TEQRKRLTIA VELVANPSII FMDEPTSGLD ARAAAIVMRA VRNTVDTGRT VVCTIHQPSI1080
1081 DIFEAFDELM LMKRGGQVIY AGPLGQNSHK VVEYFESFPG VSKIPEKYNP ATWMLEASSL1140
1141 AAELKLSVDF AELYNQSALH QRNKALVKEL SVPPAGASDL YFATQFSQNT WGQFKSCLWK1200
1201 QWWTYWRSPD YNLVRFIFTL ATSLLIGTVF WQIGGNRSNA GDLTMVIGAL YAAIIFVGIN1260
1261 NCSTVQPMVA VERTVFYRER AAGMYSAMPY AISQVTCELP YVLIQTVYYS LIVYAMVGFE1320
1321 WKAEKFFWFV FVSYFSFLYW TYYGMMTVSL TPNQQVASIF ASAFYGIFNL FSGFFIPRPK1380
1381 IPKWWIWYYW ICPVAWTVYG LIVSQYGDVE TRIQVLGGAP DLTVKQYIED HYGFQSDFMG1440
1441 PVAAVLIAFT VFFAFIFAFC IRTLNFQTR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
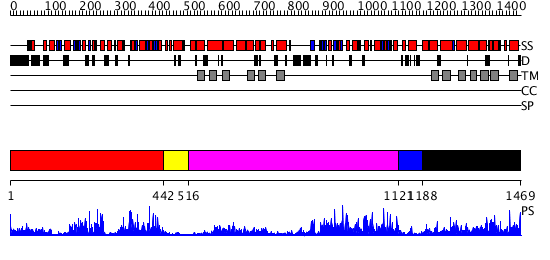
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..441] | 28.69897 | No description for 3bk7A was found. | |
| 2 | View Details | [442..515] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [516..1120] | 88.154902 | No description for 2hydA was found. | |
| 4 | View Details | [1121..1187] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1188..1469] | 1.336996 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|