













| General Information: |
|
| Name(s) found: |
ABI4_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 328 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS]
|
| Biological Process: |
sugar mediated signaling pathway
[TAS]
response to osmotic stress [IMP] regulation of transcription, DNA-dependent [ISS][TAS] seed development [IMP] hexokinase-dependent signaling [NAS] starch catabolic process [NAS] response to water deprivation [IMP] response to sucrose stimulus [IEP] abscisic acid mediated signaling pathway [IMP] response to glucose stimulus [IMP] response to abscisic acid stimulus [NAS] triglyceride catabolic process [IMP] mitochondria-nucleus signaling pathway [IDA] response to trehalose stimulus [IMP] |
| Molecular Function: |
DNA binding
[IDA][ISS]
transcription factor activity [ISS][TAS] promoter binding [IDA] transcription activator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDPLASQHQH NHLEDNNQTL THNNPQSDST TDSSTSSAQR KRKGKGGPDN SKFRYRGVRQ 60
61 RSWGKWVAEI REPRKRTRKW LGTFATAEDA ARAYDRAAVY LYGSRAQLNL TPSSPSSVSS 120
121 SSSSVSAASS PSTSSSSTQT LRPLLPRPAA ATVGGGANFG PYGIPFNNNI FLNGGTSMLC 180
181 PSYGFFPQQQ QQQNQMVQMG QFQHQQYQNL HSNTNNNKIS DIELTDVPVT NSTSFHHEVA 240
241 LGQEQGGSGC NNNSSMEDLN SLAGSVGSSL SITHPPPLVD PVCSMGLDPG YMVGDGSSTI 300
301 WPFGGEEEYS HNWGSIWDFI DPILGEFY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
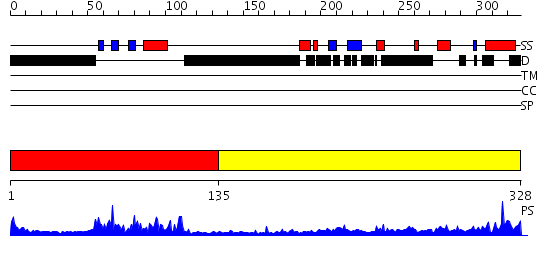
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | 25.09691 | GCC-box binding domain | |
| 2 | View Details | [135..328] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)