













| General Information: |
|
| Name(s) found: |
ADO1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 609 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][TAS]
cytosol [IDA] SCF ubiquitin ligase complex [IPI] |
| Biological Process: |
flower development
[IMP]
entrainment of circadian clock by photoperiod [IMP] SCF-dependent proteasomal ubiquitin-dependent protein catabolic process [IC] circadian rhythm [IMP][ISS][TAS] regulation of circadian rhythm [IMP] response to red light [IMP] ubiquitin-dependent protein catabolic process [TAS] proteasomal protein catabolic process [IMP] |
| Molecular Function: |
ubiquitin-protein ligase activity
[ISS]
protein binding [IPI] blue light photoreceptor activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEWDSGSDLS ADDASSLADD EEGGLFPGGG PIPYPVGNLL HTAPCGFVVT DAVEPDQPII 60
61 YVNTVFEMVT GYRAEEVLGG NCRFLQCRGP FAKRRHPLVD SMVVSEIRKC IDEGIEFQGE 120
121 LLNFRKDGSP LMNRLRLTPI YGDDDTITHI IGIQFFIETD IDLGPVLGSS TKEKSIDGIY 180
181 SALAAGERNV SRGMCGLFQL SDEVVSMKIL SRLTPRDVAS VSSVCRRLYV LTKNEDLWRR 240
241 VCQNAWGSET TRVLETVPGA KRLGWGRLAR ELTTLEAAAW RKLSVGGSVE PSRCNFSACA 300
301 VGNRVVLFGG EGVNMQPMND TFVLDLNSDY PEWQHVKVSS PPPGRWGHTL TCVNGSNLVV 360
361 FGGCGQQGLL NDVFVLNLDA KPPTWREISG LAPPLPRSWH SSCTLDGTKL IVSGGCADSG 420
421 VLLSDTFLLD LSIEKPVWRE IPAAWTPPSR LGHTLSVYGG RKILMFGGLA KSGPLKFRSS 480
481 DVFTMDLSEE EPCWRCVTGS GMPGAGNPGG VAPPPRLDHV AVNLPGGRIL IFGGSVAGLH 540
541 SASQLYLLDP TEDKPTWRIL NIPGRPPRFA WGHGTCVVGG TRAIVLGGQT GEEWMLSELH 600
601 ELSLASYLT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
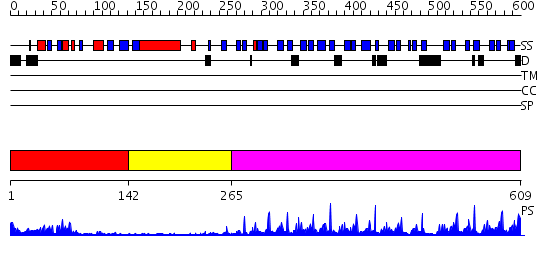
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..141] | 7.30103 | No description for 2pr5A was found. | |
| 2 | View Details | [142..264] | 2.74 | No description for 2e31A was found. | |
| 3 | View Details | [265..609] | 52.39794 | 1.35 angstrom structure of the Kelch domain of Keap1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)