













| General Information: |
|
| Name(s) found: |
RHM2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 667 amino acids |
Gene Ontology: |
|
| Cellular Component: |
soluble fraction
[IDA]
|
| Biological Process: |
UDP-rhamnose biosynthetic process
[IDA]
mucilage biosynthetic process [IMP] metabolic process [ISS] seed coat development [IMP] |
| Molecular Function: |
UDP-glucose 4,6-dehydratase activity
[IDA]
UDP-L-rhamnose synthase activity [IDA][ISS] catalytic activity [ISS] UDP-4-keto-rhamnose-4-keto-reductase activity [TAS] UDP-4-keto-6-deoxy-glucose-3,5-epimerase activity [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDDTTYKPKN ILITGAAGFI ASHVANRLIR NYPDYKIVVL DKLDYCSDLK NLDPSFSSPN 60
61 FKFVKGDIAS DDLVNYLLIT ENIDTIMHFA AQTHVDNSFG NSFEFTKNNI YGTHVLLEAC 120
121 KVTGQIRRFI HVSTDEVYGE TDEDAAVGNH EASQLLPTNP YSATKAGAEM LVMAYGRSYG 180
181 LPVITTRGNN VYGPNQFPEK MIPKFILLAM SGKPLPIHGD GSNVRSYLYC EDVAEAFEVV 240
241 LHKGEIGHVY NVGTKRERRV IDVARDICKL FGKDPESSIQ FVENRPFNDQ RYFLDDQKLK 300
301 KLGWQERTNW EDGLKKTMDW YTQNPEWWGD VSGALLPHPR MLMMPGGRLS DGSSEKKDVS 360
361 SNTVQTFTVV TPKNGDSGDK ASLKFLIYGK TGWLGGLLGK LCEKQGITYE YGKGRLEDRA 420
421 SLVADIRSIK PTHVFNAAGL TGRPNVDWCE SHKPETIRVN VAGTLTLADV CRENDLLMMN 480
481 FATGCIFEYD ATHPEGSGIG FKEEDKPNFF GSFYSKTKAM VEELLREFDN VCTLRVRMPI 540
541 SSDLNNPRNF ITKISRYNKV VDIPNSMTVL DELLPISIEM AKRNLRGIWN FTNPGVVSHN 600
601 EILEMYKNYI EPGFKWSNFT VEEQAKVIVA ARSNNEMDGS KLSKEFPEML SIKESLLKYV 660
661 FEPNKRT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
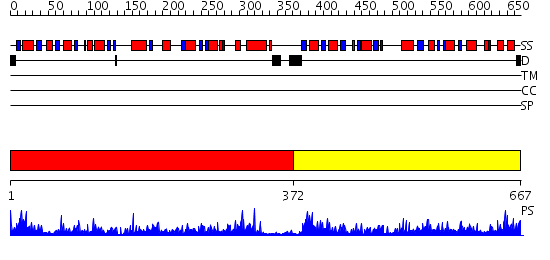
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..371] | 98.522879 | dTDP-glucose 4,6-dehydratase (RmlB) | |
| 2 | View Details | [372..667] | 45.39794 | gdp-mannose-3', 5' -epimerase (arabidopsis thaliana), with gdp-alpha-d-mannose and gdp-beta-l-galactose bound in the active site. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)