













| General Information: |
|
| Name(s) found: |
PI5K6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 715 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
phosphatidylinositol metabolic process
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
phosphatidylinositol phosphate kinase activity [IEA] 1-phosphatidylinositol-4-phosphate 5-kinase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVAHADDAD DYSRPTGESY HAEKALPSGD FYTGQWRDNL PHGHGKYLWT DGCMYVGDWH 60
61 RGKTMGKGRF SWPSGATYEG DFKNGYMDGK GTYIDSSGDL YRGSWVMNLR HGQGTKSYVN 120
121 GDCYDGEWRR GLQDGHGRYQ WKNENHYIGQ WKNGLMNGNG TMIWSNGNRY DGSWEDGAPK 180
181 GNGTFRWSDG SFYVGVWSKD PKEQNGTYYP STSSGNFDWQ PQQVFYVDLS ECVVCTCQRI 240
241 PVLPSQKMPV WYGASEQSSS GNRTKNSERP RRRSVDGRVS NGEMELRSNG SGYLQVDDNA 300
301 ESTRSSLGPL RIQPAKKQGQ TISKGHKNYE LMLNLQLGIR HSVGRPAPAT SLDLKASAFD 360
361 PKEKLWTKFP SEGSKYTPPH QSCEFKWKDY CPVVFRTLRK LFSVDAADYM LSICGNDALR 420
421 ELSSPGKSGS FFYLTNDDRY MIKTMKKAET KVLIRMLPAY YNHVRACENT LVTKFFGLHC 480
481 VKLTGTAQKK VRFVIMGNLF CTGHSIHRRF DLKGSSHGRL TTKPESEIDP NTTLKDLDLN 540
541 FAFRLQKNWF QEFCRQVDRD CEFLEQERIM DYSLLVGLHF REAAIKDSAT PTSGARTPTG 600
601 NSETRLSRAE MDRFLLDASK LASIKLGINM PARVERTARR SDCENQLVGD PTGEFYDVIV 660
661 YFGIIDILQD YDISKKLEHA YKSMQYDPTS ISAVDPKQYS RRFRDFIFRV FVEDA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
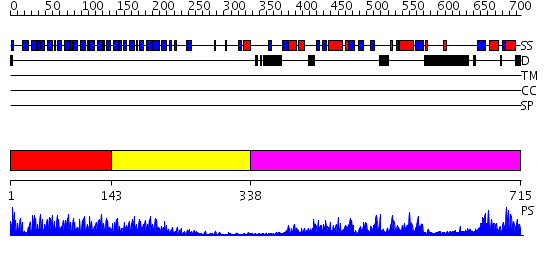
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..142] | 10.0 | Crystal structure of the complete modular teichioic acid phosphorylcholine esterase Pce (CbpE) from Streptococcus pneumoniae | |
| 2 | View Details | [143..337] | 21.30103 | Histone H3 K4-specific methyltransferase SET7/9 N-terminal domain; Histone H3 K4-specific methyltransferase SET7/9 catalytic domain | |
| 3 | View Details | [338..715] | 98.0 | Phosphatidylinositol phosphate kinase IIbeta, PIPK IIbeta |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)