













| General Information: |
|
| Name(s) found: |
LHW_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 650 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
root development
[IMP]
stele development [IMP] regulation of transcription [TAS] maintenance of root meristem identity [IMP] |
| Molecular Function: |
transcription factor activity
[ISS]
protein homodimerization activity [IPI] transcription activator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGVLLREALR SMCVNNQWSY AVFWKIGCQN SSLLIWEECY NETESSSNPR RLCGLGVDTQ 60
61 GNEKVQLLTN RMMLNNRIIL VGEGLVGRAA FTGHHQWILA NSFNRDVHPP EVINEMLLQF 120
121 SAGIQTVAVF PVVPHGVVQL GSSLPIMENL GFVNDVKGLI LQLGCVPGAL LSENYRTYEP 180
181 AADFIGVPVS RIIPSQGHKI LQSSAFVAET SKQHFNSTGS SDHQMVEESP CNLVDEHEGG 240
241 WQSTTGFLTA GEVAVPSNPD AWLNQNFSCM SNVDAAEQQQ IPCEDISSKR SLGSDDLFDM 300
301 LGLDDKNKGC DNSWGVSQMR TEVLTRELSD FRIIQEMDPE FGSSGYELSG TDHLLDAVVS 360
361 GACSSTKQIS DETSESCKTT LTKVSNSSVT TPSHSSPQGS QLFEKKHGQP LGPSSVYGSQ 420
421 ISSWVEQAHS LKREGSPRMV NKNETAKPAN NRKRLKPGEN PRPRPKDRQM IQDRVKELRE 480
481 IIPNGAKCSI DALLERTIKH MLFLQNVSKH SDKLKQTGES KIMKEDGGGA TWAFEVGSKS 540
541 MVCPIVVEDI NPPRIFQVEM LCEQRGFFLE IADWIRSLGL TILKGVIETR VDKIWARFTV 600
601 EASRDVTRME IFMQLVNILE QTMKCGGNSK TILDGIKATM PLPVTGGCSM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
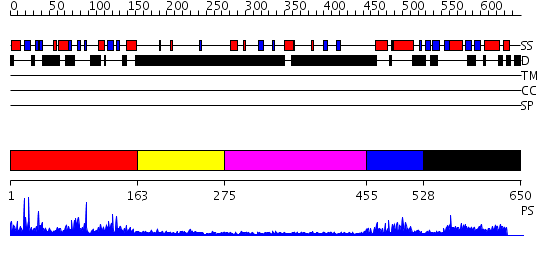
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..162] | 8.153982 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [163..274] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [275..454] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [455..527] | 12.522879 | SREBP-1a | |
| 5 | View Details | [528..650] | 2.441995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 |
|
||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)