













| General Information: |
|
| Name(s) found: |
CLSY1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1256 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA]
|
| Biological Process: |
gene silencing by RNA
[IMP]
|
| Molecular Function: |
nucleic acid binding
[IEA]
DNA binding [IEA][ISS] ATP binding [IEA][ISS] helicase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRKHYFEFN HPFNPCPFEV FCWGTWKAVE YLRIENGTMT MRLLENGQVL DDIKPFQRLR 60
61 IRSRKATLID CTSFLRPGID VCVLYQRDEE TPEPVWVDAR VLSIERKPHE SECLCTFHVS 120
121 VYIDQGCIGL EKHRMNKVPV LVGLNEIAIL QKFCKEQSLD RYYRWRYSED CSSLVKTRLN 180
181 LGKFLPDLTW LLVTSVLKNI VFQIRTVHEK MVYQIVTDED CEGSSSSLSA MNITVEDGVV 240
241 MSKVVLFNPA EDTCQDSDVK EEIEEEVMEL RRSKRRSGRP ERYGDSEIQP DSKDGWVRMM 300
301 PYRYNIWNVS SDDDDEEEDC EDDKDTDDDL YLPLSHLLRK KGSKKGFSKD KQREIVLVDK 360
361 TERKKRKKTE GFSRSCELSV IPFTPVFEPI PLEQFGLNAN SLCGGVSGNL MDEIDKYRSK 420
421 AAKYGKKKKK KIEMEEMESD LGWNGPIGNV VHKRNGPHSR IRSVSRETGV SEEPQIYKKR 480
481 TLSAGAYNKL IDSYMSRIDS TIAAKDKATN VVEQWQGLKN PASFSIEAEE RLSEEEEDDG 540
541 ETSENEILWR EMELCLASSY ILDDHEVRVD NEAFHKATCD CEHDYELNEE IGMCCRLCGH 600
601 VGTEIKHVSA PFARHKKWTT ETKQINEDDI NTTIVNQDGV ESHTFTIPVA SSDMPSAEES 660
661 DNVWSLIPQL KRKLHLHQKK AFEFLWKNLA GSVVPAMMDP SSDKIGGCVV SHTPGAGKTF 720
721 LIIAFLASYL KIFPGKRPLV LAPKTTLYTW YKEFIKWEIP VPVHLLHGRR TYCMSKEKTI 780
781 QFEGIPKPSQ DVMHVLDCLD KIQKWHAQPS VLVMGYTSFL TLMREDSKFA HRKYMAKVLR 840
841 ESPGLLVLDE GHNPRSTKSR LRKALMKVDT DLRILLSGTL FQNNFCEYFN TLCLARPKFV 900
901 HEVLVELDKK FQTNQAEQKA PHLLENRARK FFLDIIAKKI DTKVGDERLQ GLNMLRNMTS 960
961 GFIDNYEGSG SGSGDVLPGL QIYTLLMNST DVQHKSLTKL QNIMSTYHGY PLELELLITL1020
1021 AAIHPWLVKT TTCCAKFFNP QELLEIEKLK HDAKKGSKVM FVLNLVFRVV KREKILIFCH1080
1081 NIAPIRLFLE LFENVFRWKR GRELLTLTGD LELFERGRVI DKFEEPGGQS RVLLASITAC1140
1141 AEGISLTAAS RVIMLDSEWN PSKTKQAIAR AFRPGQQKVV YVYQLLSRGT LEEDKYRRTT1200
1201 WKEWVSSMIF SEEFVEDPSQ WQAEKIEDDV LREIVEEDKV KSFHMIMKNE KASTGG |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
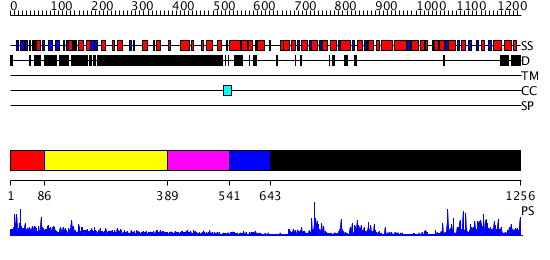
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [86..388] | 7.0 | Heavy meromyosin subfragment | |
| 3 | View Details | [389..540] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [541..642] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [643..1256] | 73.0 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)