













| General Information: |
|
| Name(s) found: |
DEXH4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1417 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
nucleic acid binding
[IEA][ISS]
ATP binding [IEA][ISS] helicase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPTKKPQKN KQSKNEIASS LIPNSGHKKP SKAPKLLISP ENEDRLRRLL LNFRRTPSPV 60
61 TATLSVTQKR KKLNNLYENL SCEGFLDNQI ELVLSSLRDG ATLETALDWL CLNLPSHELP 120
121 VNFSNGASRF PSTGRSVAVI SKSKKDWNVS AESSVQEVKE VPESEVLVRV KSKRDEEEED 180
181 SLSSCQPSQA DWIHQYMKRL EEEELESSDD ERDKVSGPRS FEVIAKEYCV ERYNAIKAKR 240
241 KGDKSGQSQA GLAICKLKEE MNALGPSEAI LESEFQRDRQ DCEGAREKEV ASSVLDDVHE 300
301 SVNADAFFFQ LFDDLTLDTN PVGSCKSEEQ ESVVSNDSLD DLDLGDLFDE DVPPYVPSPH 360
361 ELLELQKKEI MRELCNEKHL TKLNGIWKKG EAQKIPKALL HQLCQRSGWI APKFNKVTGE 420
421 GRNFSYTTSV MRKSSGFGKS RQAGGLVTIQ LPHQVEDFES IQDAQNRVAA FALHKLFSDL 480
481 PVHFAITEPY ASLVLIWKQE ESLGITSREE QRREKFVESL LEADNFSLTT TSRGIHSALP 540
541 MVDSCVKEND DLDVVKSNHR ARRNSSMAAE CSSLKQKQEN KKKMQKYKDM LKTRAALPIS 600
601 EVKKDILQKL KEKDVLVVCG ETGSGKTTQV PQFILDDMID SGHGGYCNII CTQPRAITVA 660
661 QRVADERCEP PPGFDNSVVA YQVRHQNARS DKTRLLFCTT GILLRKLVGD TTLKDVTHII 720
721 VDEVHERSLM GDFLLIILKS LIEKQSWDNA LPKLKVILMS ATVDAHQFSR YFGQCPIITA 780
781 QGRTHPVTTY FLEDIYERTK YLLASDSPAA LSSDTSITDK LGSVNVPRGK KNLMLAGWGD 840
841 SYLVSEDSLN TSYDSIKYIA SAVVDYDLLE ELICHIDDTC EEGAILVFLP GMSEINMLLN 900
901 RLAASYRFRG ASGDWLLPLH SSIASTEQKK VFLRPPKGIR KVIIATNIAE TSITIEDVVY 960
961 VIDSGKHKEN RYNPHKKLSS MVEDWVSKAN ARQRMGRAGR VKPGHCFSLY TRHRFEKLMR1020
1021 PYQVPEMLRV PLVELCLHIK LLGLGQIKPF LSKALEPPSE SAINSAILLL HKVGALEGDE1080
1081 ELTPLGHHLA KLPVDLLIGK MLLYGGIFGC LSPILSIAAF LSCCKSPFVY AKDEQNVDRV1140
1141 KLALLSDKLE SSSNLNNNDR QSDHLLMVVA YEKWVRILHE QGFKAAESFC ESKFLNSSVM1200
1201 RMMRERRVEF GMLLADIGLI NLPKGKGRRK ENFDVWFSDK TQPFNMYSQE PEVVKAILCA1260
1261 GLCPNIAEGL VNRLTKPAEE TQRYAVWHDG KREVHIHRNS INKNCKAFQY PFIVFLEKLE1320
1321 TKKVVYLQDT TVVSPFSILL FGGSINVHHQ SGSVTIDGWL KLTAPAQTAV LFKELRLTLH1380
1381 SILKDLIRKP EKSGIVHNEV VKSMVHLLIE EGKPQHT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
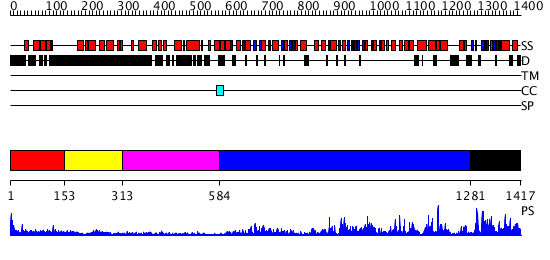
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..152] | 1.287995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [153..312] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [313..583] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [584..1280] | 16.69897 | No description for 2p6rA was found. | |
| 5 | View Details | [1281..1417] | 2.593976 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)