













| General Information: |
|
| Name(s) found: |
VPS9A_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 520 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
membrane [IDA] cytosol [IDA] |
| Biological Process: |
cell plate assembly
[IMP]
embryonic development [IMP] transport [ISS] post-embryonic root development [IMP] cell wall biogenesis [IMP] |
| Molecular Function: |
Rho guanyl-nucleotide exchange factor activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MENTDVFLGL HDFLERMRKP SAGDFVKSIK SFIVSFSNNA PDPEKDCAMV QEFFSKMEAA 60
61 FRAHPLWSGC SEEELDSAGD GLEKYVMTKL FTRVFASNTE EVIADEKLFQ KMSLVQQFIS 120
121 PENLDIQPTF QNESSWLLAQ KELQKINMYK APRDKLVCIL NCCKVINNLL LNASIASNEN 180
181 APGADEFLPV LIYVTIKANP PQLHSNLLYI QRYRRESKLV GEAAYFFTNI LSAESFISNI 240
241 DAKSISLDEA EFEKNMESAR ARISGLDSQT YQTGHGSAPP PRDESTLQKT QSLNPKRENT 300
301 LFQSKSSDSL SGTNELLNIN SETPMKKAES ISDLENKGAT LLKDTEPSKV FQEYPYIFAS 360
361 AGDLRIGDVE GLLNSYKQLV FKYVCLTKGL GDGTSLAPSS SPLQASSGFN TSKESEDHRR 420
421 SSSDVQMTKE TDRSVDDLIR ALHGEGEDVN NLSDVKHEEY GAMLVEGKDE ERDSKVQGEV 480
481 DAKDIELMKQ IPKREGDNSS SRPAEDEDVG SKQPVTEASE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
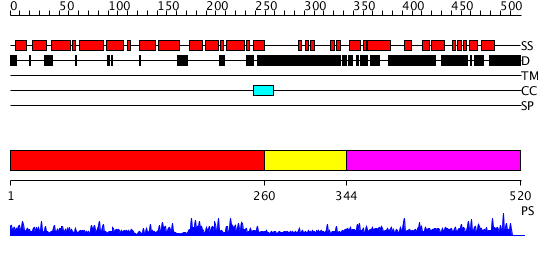
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..259] | 70.30103 | Crystal Structure of the Vps9 Domain of Rabex-5 | |
| 2 | View Details | [260..343] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [344..520] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)