













| General Information: |
|
| Name(s) found: |
gi|6996266
[NCBI NR]
gi|15231193 [NCBI NR] gi|11357925 [NCBI NR] |
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 989 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA]
nucleotide binding [IEA] zinc ion binding [IEA] RNA binding [ISS] protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDYGEKTCP LCAEEMDLTD QQLKPCKCGY QICVWCWHHI MDMAEKDQSE GRCPACRTPY 60
61 DKEKIVGMTV DQERLASEGN MDRKKIQKSK PKSSDGRKPL TSVRVVQRNL VYIVGLPLNL 120
121 ADEDLLQRKE YFGQYGKVLK VSMSRTATGL IQQFPNNTCS VYITYGKEEE AIRCIQSVHG 180
181 FILDGKALKA CFGTTKYCHA WLRNVACNNQ DCLYLHEVGS QEDSFTKDEI ISAHTRVQQI 240
241 TGATNTMQYR SGSMLPPPLD AYTSDSSTGN PIAKVPSSTS VSAPKSSPPS GSSGKSTALP 300
301 AAASWGARLT NQHSLATSAL SNGSLDNQRS TSENGTLATS TVVTKAANGP VSSSNSLQKA 360
361 PLKEEIQSLA EKSKPGVLKP LQQKIVLDPE SKRTTSPNRD PSSNQISCLV ESSYNSRVID 420
421 KPSAVENSLE HTSEIAEDVF DVGKLSADVA WMGITTNSRD ETPGVPVVIG THCDLGSITQ 480
481 SDNDVQNLEQ CRKQSPTNTY AEADISLNGI HGSRPEWDWR SGLQSQIDVK EPLEVNDFSS 540
541 FNNNRRGIAE AVSHSTSKFS SSISILDSNH LASRSFQNRE TSCGMDSKTG SSFEIGSDRL 600
601 HLPNGFSEKA MSNMEHSLFA NEGRSNIQNT EDDIISNILD FDPWDESLTS QHNFAKLLGQ 660
661 SDHRASTLES SNLLKQHNDQ SRFSFARHEE SNSQAYDNRS YSIYGQLSRD QPLQEFGANR 720
721 DMYQDKLGSQ NGFASNYSGG YEQFATSPGL SSYKSPVART QVSAPPGFSA PNRLPPPGFS 780
781 SHQRGDLSSD IASGTRLLDS ANLLRNAYHV PPPSGNLNAA GDIEFIDPAI LAVGRGRLHN 840
841 GMETADFDLR SGFSSQLNSF DNDARLQLLA QRSLAAQQVN GFHDPRNVNN FSSSFSDPYG 900
901 ISSRPTDQTQ GTGLSPFTQL PRQASANPLL SNGHWDNKWN EPQSGNNLGI TQLLRNERMG 960
961 FNDNVYSGFE EPKFRRPGPG DPYNRTYGI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
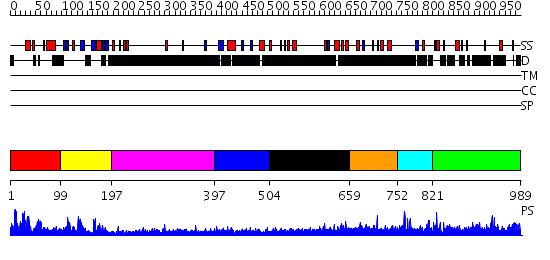
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | 32.69897 | Not-4 N-terminal RING finger domain | |
| 2 | View Details | [99..196] | 27.39794 | Solution structure of the RNA recognition motif of CNOT4 | |
| 3 | View Details | [197..396] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [397..503] | 3.306992 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [504..658] | 5.286992 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [659..751] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [752..820] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [821..989] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. | ||||||
| 6 | No functions predicted. | ||||||
| 7 | No functions predicted. | ||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)