













| General Information: |
|
| Name(s) found: |
gi|25404679
[NCBI NR]
gi|238478996 [NCBI NR] gi|17065202 [NCBI NR] gi|15220477 [NCBI NR] gi|12324671 [NCBI NR] |
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 752 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA]
|
| Biological Process: |
protein amino acid phosphorylation
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
protein kinase activity [IEA] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAGRNIRYP DHELRDQESN SRFSRRDSAY ANEDYNHVRN GAIDNGKGRV SNLRHGDRDR 60
61 IKSGARQEEN KMVSSGFRLS KSNPGSREVF IDLGPKRCGF SARSVDREPG ELSSESGSDD 120
121 LIESESLAKV NGVVKEVENR AQSPVEKKRK FSPIVWDRDD HERSNLSRNE KPVEVTPLPP 180
181 PPPLVKRSSQ SPSVSCGGNS HYSPAKSDMH QDPVEVGVSA VSMPALSPSV EMSSLCVVEQ 240
241 SSNAEQDDKQ EHATHLEEDE NMPTRHISSS RWAAGNSSPT DEVEIVEEVG EKKRRKKPFP 300
301 VQGRFRNTSQ TPEVGELVRE GYRSSDSDER GHHSLPGSRD DFEERDAVKS DKMEIDEEEH 360
361 RRENSVDSLS ETDSDDEYVR HETPEPASTP LRSINMLQGC RSVDEFERLN KIDEGTYGVV 420
421 YRAKDKKTGE IVALKKVKME KEREGFPLTS LREINILLSF HHPSIVDVKE VVVGSSLDSI 480
481 FMVMEYMEHD LKALMETMKQ RFSQSEVKCL MLQLLEGVKY LHDNWVLHRD LKTSNLLLNN 540
541 RGELKICDFG LARQYGSPLK PYTHLVVTLW YRAPELLLGA KQYSTAIDMW SLGCIMAELL 600
601 MKAPLFNGKT EFDQLDKIFR ILGTPNESIW PGFSKLPGVK VNFVKHQYNL LRKKFPATSF 660
661 TGAPVLSDAG FDLLNKLLTY DPERRITVNE ALKHDWFREV PLPKSKDFMP TFPAQHAQDR 720
721 RGRRMVKSPD PLEEQRRKEL TQTELGSGGL FG |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
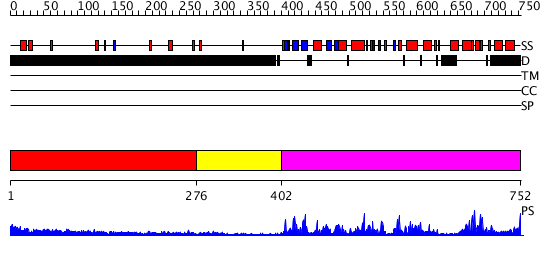
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..275] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [276..401] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [402..752] | 90.522879 | Cyclin-dependent PK, CDK2 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)