













| General Information: |
|
| Name(s) found: |
gi|9280653
[NCBI NR]
gi|240254006 [NCBI NR] gi|14532736 [NCBI NR] gi|13430452 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 740 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein amino acid phosphorylation
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
protein tyrosine kinase activity [IEA] protein kinase activity [IEA] protein serine/threonine kinase activity [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGCVNSRHRP FRRKSTTLKE SSEEKRSSRI DSSRRIDDWI QPEDGFDRLS NSGDAKVRLI 60
61 ESEMFSTSRC HDHQIGKILE NPATVAHMDR VVHDQELRRA SSAVVDSDLD IDPKVVKAKL 120
121 DRWNSKDSKV RLIESEKLSS SMFSEHHQIE KGVEKPEVEA SVRVVHRELK RGSSIVSPKD 180
181 AERKQVAAGW PSWLVSVAGE SLVDWAPRRA NTFEKLEKIG QGTYSSVYRA RDLLHNKIVA 240
241 LKKVRFDLND MESVKFMARE IIVMRRLDHP NVLKLEGLIT APVSSSLYLV FEYMDHDLLG 300
301 LSSLPGVKFT EPQVKCYMRQ LLSGLEHCHS RGVLHRDIKG SNLLIDSKGV LKIADFGLAT 360
361 FFDPAKSVSL TSHVVTLWYR PPELLLGASH YGVGVDLWST GCILGELYAG KPILPGKTEV 420
421 EQLHKIFKLC GSPTENYWRK QKLPSSAGFK TAIPYRRKVS EMFKDFPASV LSLLETLLSI 480
481 DPDHRSSADR ALESEYFKTK PFACDPSNLP KYPPSKEIDA KMRDEAKRQQ PMRAEKQERQ 540
541 DSMTRISHER KFVPPVKANN SLSMTMEKQY KDLRSRNDSF KSFKEERTPH GPVPDYQNMQ 600
601 HNRNNQTGVR ISHSGPLMSN RNMAKSTMHV KENALPRYPP ARVNPKMLSG SVSSKTLLER 660
661 QDQPVTNQRR RDRRAYNRAD TMDSRHMTAP IDPSWYNPSD SKIYMSGPLL AQPSRVDQML 720
721 EEHDRQLQEF NRQALKTPQG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
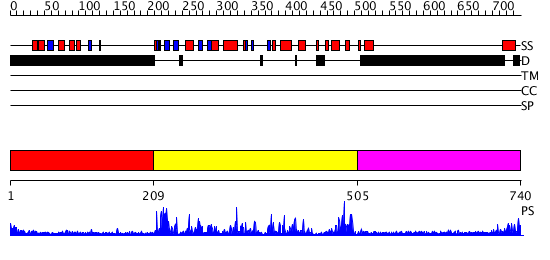
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..208] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [209..504] | 86.39794 | Structure of CDK2/Cyclin A with PNU-292137 | |
| 3 | View Details | [505..740] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)