













| General Information: |
|
| Name(s) found: |
gi|7485133
[NCBI NR]
gi|7268405 [NCBI NR] gi|2245011 [NCBI NR] gi|15235783 [NCBI NR] |
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 743 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
proteolysis
[IEA]
|
| Molecular Function: |
aspartic-type endopeptidase activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVSPVTLAPP SPEGFYAINN PFLVNGPKGF LEAKTLDNES YIRFDLPGVP RDGVTVLLDQ 60
61 PNNTVNILAD APKEHKYDSS HRNYDGTKLR PKISVGGINT KIPCKCCEFF SGFTSHMADG 120
121 VLRLVLTTSH ASTRRPSCTC ISFLGGSDGE DRVGTATDGF PHATDPNDGY YATNNPYQVN 180
181 GPKGFTEFKY LEETHDLFVR LDFPGIQKES VIILLEPSKK AVIVTGEAPK ESKHDSSHRK 240
241 YGTATGLICD CCEISNIQCF VGDGVVRLIL SKQKINLRVP IFCSCKITNL QNVLFFCLIF 300
301 VGGARMPTDA SPNIIRGYNP EDLSVELLLE TSLRCFSIVN VSDIFHGFVL LIPIVAVAGG 360
361 HPLAHLRGLN PEGCRGTDPF DPAFTGPTIR PHPSVLEGST SAYETKQLSN GGLYLRIDMP 420
421 GVPSDGFIVA VDGNGVVTIM GRAPATMHDS NGFYAMNNPY QANGPKGFAE FKYLEDTHDL 480
481 FVRLDLPGVK KETVGFYAIN NQFQANGPKG FMEFKMLENE DMYVRIDFPG VPRDGVRVFL 540
541 DQSKKAVCIF ADAPKEHKYD YSERNYGTST GLVCKCCEIS GFTSFMSDGV LRLLLTKSNI 600
601 TPQRSSCISF LAGPDFREDL RGNGPHKFPH GTDPNDPKLT GRILMPHPCV NYGSEMAYES 660
661 KQLQNGGLYV RVDMPGVPKE NFTVAVMNGR VRVTGEAPAI SHDSSGRFYT GDVAMLSTPF 720
721 DIPIRKIKII AKNGVIRLII PPV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
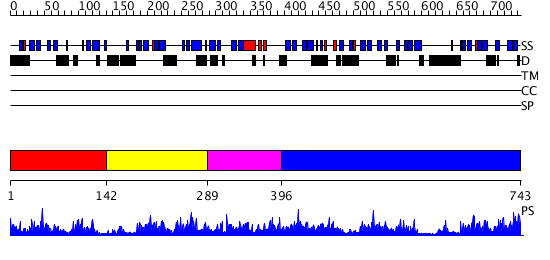
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..141] | 4.98 | Small heat shock protein | |
| 2 | View Details | [142..288] | 26.522879 | Small heat shock protein | |
| 3 | View Details | [289..395] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [396..743] | 12.221849 | CRYSTAL STRUCTURE AND ASSEMBLY OF TSP36, A METAZOAN SMALL HEAT SHOCK PROTEIN |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.70 |
Source: Reynolds et al. (2008)