













| General Information: |
|
| Name(s) found: |
Y5872_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 519 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
mismatch repair
[ISS]
|
| Molecular Function: |
ATP binding
[ISS]
damaged DNA binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKQKNQHKKK KKRSCAAKPS GDGTTSDGNK KDVEEERKDG EGKREIENVG KNFIESLMEA 60
61 FCSVSMEEAM AAYKEAGGDL NKAAEILSDL VESGDDPSTS SVASGSSGQE TASTSEYGAG 120
121 SSSSCSEDLT RDRWFKGSKQ SRVIAATGMV SSVIAKDYLK PNPVRKEFPM MERSKELCGN 180
181 GKKAADREKA EQFLSSMLGD DCELSMAVVR DVLCQCGYDV DMALNVLLDM SSSSTDDSLS 240
241 GKCFGIGVSD SLAESSFDTD TSDCELFWGG DYSQRDYAKA LMSSQDPFAT TQGIDELGLP 300
301 QKVLESLFNI RQNPKHESKT TSWRNVAKKM QSLGIDASSS SGEEPHPNTF VKDDSYHELR 360
361 KGANDQWNVT KSYYQKAAEA YSKGGRAHAA YLSDKGRVAS KQAQRADERA SQDIFVARNK 420
421 GIENVVTIDL HGQHVKPAMK LLKLHLLFGS YVPSIQTLRV ITGCGASGFG KSKVKQSVVK 480
481 LLEREGVRYC EENRGTLLIK LDGGSREFSF LDTESDSDE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
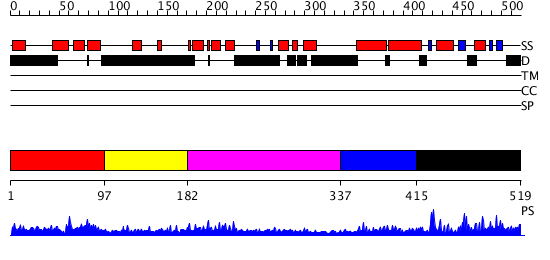
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | 0.95 | No description for 2daiA was found. | |
| 2 | View Details | [97..181] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [182..336] | 1.118996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [337..414] | 21.366532 | No description for PF08590.1 was found. Confident ab initio structure predictions are available. | |
| 5 | View Details | [415..519] | 20.69897 | No description for 2d9iA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)