













| General Information: |
|
| Name(s) found: |
URT1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 764 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
nucleotidyltransferase activity
[IEA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADGGAEPPA PPSSINAGEF LLSILHGSPS PSSQGPQHHQ SFALDPAIAA IGPTVNNPFP 60
61 PSNWQSNGHR PSNHNPPSWP LAFSPPHNLS PNFLGFPQFP PSPFTTNQFD GNQRVSPEDA 120
121 YRLGFPGTTN PAIQSMVQQQ QQQQLPPPQS ETRKLVFGSF SGDATQSLNG LHNGNLKYDS 180
181 NQHEQLMRHP QSTLSNSNMD PNLSHHRNHD LHEQRGGHSG RGNWGHIGNN GRGLKSTPPP 240
241 PPPGFSSNQR GWDMSLGSKD DDRGMGRNHD QAMGEHSKVW NQSVDFSAEA NRLRGLSIQN 300
301 ESKFNLSQQI DHPGPPKGAS LHSVSAADAA DSFSMLNKEA RRGGERREEL GQLSKAKREG 360
361 NANSDEIEDF GEDIVKSLLL EDETGEKDAN DGKKDSKTSR EKESRVDNRG QRLLGQKARM 420
421 VKMYMACRND IHRYDATFIA IYKSLIPAEE ELEKQRQLMA HLENLVAKEW PHAKLYLYGS 480
481 CANSFGFPKS DIDVCLAIEG DDINKSEMLL KLAEILESDN LQNVQALTRA RVPIVKLMDP 540
541 VTGISCDICI NNVLAVVNTK LLRDYAQIDV RLRQLAFIVK HWAKSRRVNE TYQGTLSSYA 600
601 YVLMCIHFLQ QRRPPILPCL QEMEPTYSVR VDNIRCTYFD NVDRLRNFGS NNRETIAELV 660
661 WGFFNYWAYA HDYAYNVVSV RTGSILGKRE KDWTRRVGND RHLICIEDPF ETSHDLGRVV 720
721 DKFSIRVLRE EFERAARIMH QDPNPCAKLL EPYIPEDNNG QGHN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
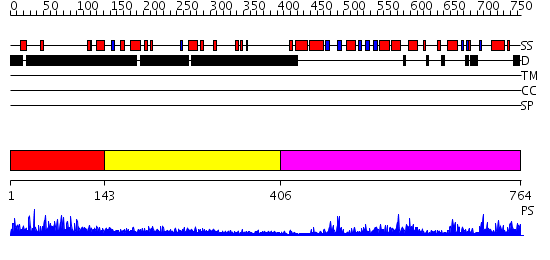
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..142] | 12.143997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [143..405] | 1.26198 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [406..764] | 53.0 | Poly(A) polymerase, middle domain; Poly(A) polymerase N-terminal, catalytic domain; Poly(A) polymerase, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)