













| General Information: |
|
| Name(s) found: |
gi|4512682
[NCBI NR]
gi|25370821 [NCBI NR] gi|20198081 [NCBI NR] gi|15227985 [NCBI NR] |
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 788 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
vesicle-mediated transport
[IEA]
vesicle docking during exocytosis [IEA] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALIDVAISC LNSIREIEED VKDAIVYIDA GCTESFQFVG AFPLFLELGA RAVCSLENMT 60
61 SLDAVADWNS KSDCAKRIVI MTSRLLNDAH RYMLRCLSTH EGPDAYREYE TLLVQDYNEH 120
121 TKKSDKISKD KGVSKFSSAL ESLTMEPIES ENVDISSGGA QGSVAEASLS RQHEDSLSFG 180
181 LPPISTGSMS DTDDVPPGAT LTAHFLYQLA LKMELKLEIF SLGDQSKNVG KILTDMSSVY 240
241 DVARRKRSAG LLLVDRTLDL ITPCCHGDSL FDRIFSSLPR AERFSSQAQL KQGVPSINRP 300
301 SLDVQVPLGE LLNEEPSKIR DSGLPEGIEA FLRGWDSYTS APQNVGLFNE CDKKSTTNWT 360
361 ELLNGSLVAT ECFRGTPYLE AMIDRKTKDG SVLVKKWLQE ALRRENISVN VRARPGYATK 420
421 PELQAMIKAL SQSQSSLLKN KGIIQLGAAT AAALDESQSA KWDTFSSAEM MLNVSAGDTS 480
481 QGLAAQISDL INKSAVAELQ AKKNEKPDSS SRGLLSFRDA LLLTIVGYIL AGENFPTSGS 540
541 GGPFSWQEEH FLKEAIVDAV LENPSAGNLK FLNGLTEELE GRLNRLKSEE TKEIPSDDQL 600
601 DIDALDDDPW GKWGDEEEEE VDNSKADESY DDMQLKLDLR DRVDSLFRFL HKLSSLRTRN 660
661 LPLREGSLAS ESSFPGEPSG NKGLVYRLIT KVLSKQEIPG LEYHSSTVGR FIKSGFGRFG 720
721 LGQAKPSLAD QSVILVFVIG GINGIEVLEA QEAVSESGRP DINLVIGGTT LLTPDDMFEL 780
781 LLGQFSHF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
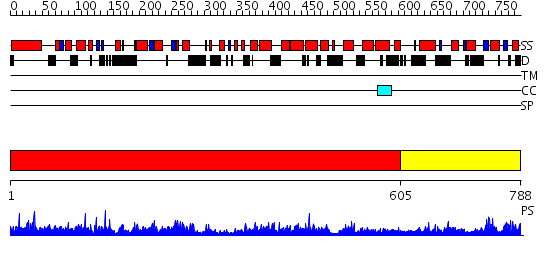
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..604] | 1.18 | Neuronal Sec1, NSec1 | |
| 2 | View Details | [605..788] | 34.69897 | Neuronal Sec1, NSec1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)