













| General Information: |
|
| Name(s) found: |
MB3R1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 776 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
regulation of transcription, DNA-dependent
[ISS]
regulation of transcription [IDA] |
| Molecular Function: |
DNA binding
[TAS]
transcription factor activity [ISS][TAS] transcription coactivator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKREMKAPTT PLESLQGDLK GKQGRTSGPA RRSTKGQWTP EEDEVLCKAV ERFQGKNWKK 60
61 IAECFKDRTD VQCLHRWQKV LNPELVKGPW SKEEDNTIID LVEKYGPKKW STISQHLPGR 120
121 IGKQCRERWH NHLNPGINKN AWTQEEELTL IRAHQIYGNK WAELMKFLPG RSDNSIKNHW 180
181 NSSVKKKLDS YYASGLLDQC QSSPLIALQN KSIASSSSWM HSNGDEGSSR PGVDAEESEC 240
241 SQASTVFSQS TNDLQDEVQR GNEEYYMPEF HSGTEQQISN AASHAEPYYP SFKDVKIVVP 300
301 EISCETECSK KFQNLNCSHE LRTTTATEDQ LPGVSNDAKQ DRGLELLTHN MDNGGKNQAL 360
361 QQDFQSSVRL SDQPFLSNSD TDPEAQTLIT DEECCRVLFP DNMKDSSTSS GEQGRNMVDP 420
421 QNGKGSLCSQ AAETHAHETG KVPALPWHPS SSEGLAGHNC VPLLDSDLKD SLLPRNDSNA 480
481 PIQGCRLFGA TELECKTDTN DGFIDTYGHV TSHGNDDNGG FPEQQGLSYI PKDSLKLVPL 540
541 NSFSSPSRVN KIYFPIDDKP AEKDKGALCY EPPRFPSADI PFFSCDLVPS NSDLRQEYSP 600
601 FGIRQLMISS MNCTTPLRLW DSPCHDRSPD VMLNDTAKSF SGAPSILKKR HRDLLSPVLD 660
661 RRKDKKLKRA ATSSLANDFS RLDVMLDEGD DCMTSRPSES PEDKNICASP SIARDNRNCA 720
721 SARLYQEMIP IDEEPKETLE SGGVTSMQNE NGCNDGGASA KNVSPSLSLH IIWYQL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
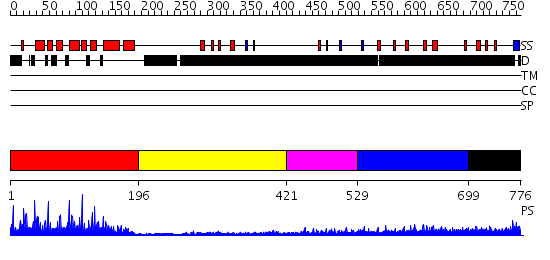
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..195] | 70.39794 | c-Myb, DNA-binding domain | |
| 2 | View Details | [196..420] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [421..528] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [529..698] | 4.169997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [699..776] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)