













| General Information: |
|
| Name(s) found: |
gi|94450858
[NCBI NR]
gi|7594553 [NCBI NR] gi|15229816 [NCBI NR] gi|11358029 [NCBI NR] |
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1531 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
nucleolus [IDA] |
| Biological Process: |
[NOT]production of miRNAs involved in gene silencing by miRNA
[IGI]
RNA processing [ISS] maintenance of DNA methylation [IMP] production of siRNA involved in RNA interference [IMP] production of ta-siRNAs involved in RNA interference [IMP] |
| Molecular Function: |
RNA binding
[ISS]
ribonuclease III activity [ISS] double-stranded RNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHSSLEPEKM EEGGGSNSLK RKFSEIDGDQ NLDSVSSPMM TDSNGSYELK VYEVAKNRNI 60
61 IAVLGTGIDK SEITKRLIKA MGSSDTDKRL IIFLAPTVNL QCCEIRALVN LKVEEYFGAK 120
121 GVDKWTSQRW DEEFSKHDVL VMTPQILLDV LRSAFLKLEM VCLLIIDECH HTTGNHPYAK 180
181 LMKIFNPEER EGVEKFATTV KEGPILYNPS PSCSLELKEK LETSHLKFDA SLRRLQELGK 240
241 DSFLNMDNKF ETYQKRLSID YREILHCLDN LGLICAHLAA EVCLEKISDT KEESETYKEC 300
301 SMVCKEFLED ILSTIGVYLP QDDKSLVDLQ QNHLSAVISG HVSPKLKELF HLLDSFRGDK 360
361 QKQCLILVER IITAKVIERF VKKEASLAYL NVLYLTENNP STNVSAQKMQ IEIPDLFQHG 420
421 KVNLLFITDV VEEGFQVPDC SCMVCFDLPK TMCSYSQSQK HAKQSNSKSI MFLERGNPKQ 480
481 RDHLHDLMRR EVLIQDPEAP NLKSCPPPVK NGHGVKEIGS MVIPDSNITV SEEAASTQTM 540
541 SDPPSRNEQL PPCKKLRLDN NLLQSNGKEK VASSKSKSSS SAAGSKKRKE LHGTTCANAL 600
601 SGTWGENIDG ATFQAYKFDF CCNISGEVYS SFSLLLESTL AEDVGKVEMD LYLVRKLVKA 660
661 SVSPCGQIRL SQEELVKAKY FQQFFFNGMF GKLFVGSKSQ GTKREFLLQT DTSSLWHPAF 720
721 MFLLLPVETN DLASSATIDW SAINSCASIV EFLKKNSLLD LRDSDGNQCN TSSGQEVLLD 780
781 DKMEETNLIH FANASSDKNS LEELVVIAIH TGRIYSIVEA VSDSSAMSPF EVDASSGYAT 840
841 YAEYFNKKYG IVLAHPNQPL MKLKQSHHAH NLLVDFNEEM VVKTEPKAGN VRKRKPNIHA 900
901 HLPPELLARI DVPRAVLKSI YLLPSVMHRL ESLMLASQLR EEIDCSIDNF SISSTSILEA 960
961 VTTLTCPESF SMERLELLGD SVLKYVASCH LFLKYPDKDE GQLSRQRQSI ISNSNLHRLT1020
1021 TSRKLQGYIR NGAFEPRRWT APGQFSLFPV PCKCGIDTRE VPLDPKFFTE NMTIKIGKSC1080
1081 DMGHRWVVSK SVSDCAEALI GAYYVSGGLS ASLHMMKWLG IDVDFDPNLV VEAINRVSLR1140
1141 CYIPKEDELI ELERKIQHEF SAKFLLKEAI THSSLRESYS YERLEFLGDS VLDFLITRHL1200
1201 FNTYEQTGPG EMTDLRSACV NNENFAQVAV KNNLHTHLQR CATVLETQIN DYLMSFQKPD1260
1261 ETGRSIPSIQ GPKALGDVVE SIAGALLIDT RLDLDQVWRV FEPLLSPLVT PDKLQLPPYR1320
1321 ELNELCDSLG YFFRVKCSND GVKAQATIQL QLDDVLLTGD GSEQTNKLAL GKAASHLLTQ1380
1381 LEKRNISRKT SLGDNQSSMD VNLACNHSDR ETLTSETTEI QSIVIPFIGP INMKKGGPRG1440
1441 TLHEFCKKHL WPMPTFDTSE EKSRTPFEFI DGGEKRTSFS SFTSTITLRI PNREAVMYAG1500
1501 EARPDKKSSF DSAVVELLYE LERRKIVIIQ K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
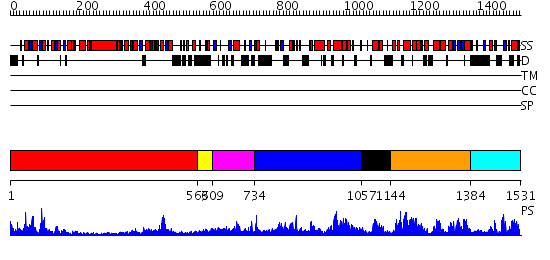
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..562] | 33.154902 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | |
| 2 | View Details | [563..608] | 35.522879 | Structure of the RecQ Catalytic Core | |
| 3 | View Details | [609..733] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [734..1056] | 45.39794 | Crystal Structure of Dicer from Giardia intestinalis | |
| 5 | View Details | [1057..1143] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1144..1383] | 51.0 | RNase III endonuclease catalytic domain; RNase III, C-terminal domain | |
| 7 | View Details | [1384..1531] | 13.045757 | dsRNA-dependent protein kinase pkr |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||
| 7 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)