













| General Information: |
|
| Name(s) found: |
RAD50_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1316 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] Mre11 complex [TAS] |
| Biological Process: |
DNA repair
[ISS]
telomere maintenance [IMP] telomere capping [IMP] mitotic recombination [IMP] double-strand break repair [IMP] |
| Molecular Function: |
ATP binding
[IEA]
zinc ion binding [IEA] nuclease activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTVDKMLIK GIRSFDPENK NVVTFFRPLT LIVGANGAGK TTIIECLKVS CTGELPPNAR 60
61 SGHSFIHDPK VAGETETKAQ IKLRFKTAAG KDVVCIRSFQ LTQKASKMEY KAIESVLQTI 120
121 NPHTGEKVCL SYRCADMDRE IPALMGVSKA ILENVIFVHQ DESNWPLQDP STLKKKFDDI 180
181 FSATRYTKAL EVIKKLHKDQ AQEIKTFKLK LENLQTLKDA AYKLRESIAQ DQERTESSKV 240
241 QMLELETSVQ KVDAEVHNKE MMLKDLRKLQ DQVSIKTAER STLFKEQQRQ YAALPEENED 300
301 TIEELKEWKS KFEERLALLG TKIRKMEREM VDTETTISSL HNAKTNYMLE ISKLQTEAEA 360
361 HMLLKNERDS TIQNIFFHYN LGNVPSTPFS TEVVLNLTNR IKSRLGELEM DLLDKKKSNE 420
421 TALSTAWDCY MDANDRWKSI EAQKRAKDEI KMGISKRIEE KEIERDSFEF EISTVDVKQT 480
481 DEREKQVQVE LERKTKQNSE RGFESKIEQK QHEIYSLEHK IKTLNRERDV MAGDAEDRVK 540
541 LSLKKTEQEN LKKKHKKIID ECKDRIRGVL KGRLPPEKDM KREIVQALRS IEREYDDLSL 600
601 KSREAEKEVN MLQMKIQEVN NSLFKHNKDT ESRKRYIESK LQALKQESVT IDAYPKLLES 660
661 AKDKRDDRKR EYNMANGMRQ MFEPFEKRAR QEHSCPCCER SFTADEEASF IKKQRVKASS 720
721 TGEHLKALAV ESSNADSVFQ QLDKLRAVFE EYSKLTTEII PLAEKTLQEH TEELGQKSEA 780
781 LDDVLGISAQ IKADKDSIEA LVQPLENADR IFQEIVSYQK QIEDLEYKLD FRGLGVKTME 840
841 EIQSELSSLQ SSKDKLHGEL EKLRDDQIYM ERDISCLQAR WHAVREEKAK AANLLRDVTK 900
901 AEEDLERLAE EKSQLDLDVK YLTEALGPLS KEKEQLLSDY NDMKIRRNQE YEELAEKKRN 960
961 YQQEVEALLK ASYKINEYHD LKKGERLDDI QEKQRLSDSQ LQSCEARKNE LAGELNRNKD1020
1021 LMRNQDQLRR NIEDNLNYRT TKAKVEELTR EIESLEEQIL NIGGIAAVEA EIVKILRERE1080
1081 RLLSELNRCR GTVSVYESSI SKNRVELKQA QYKDIDKRHF DQLIQLKTTE MANKDLDRYY1140
1141 NALDKALMRF HTMKMEEINK IIRELWQQTY RGQDMDYIRI HSDSEGAGTR SYSYKVLMQT1200
1201 GDTELEMRGR CSAGQKVLAS LIIRLALAET FCLNCGILAL DEPTTNLDGP NSESLAGALL1260
1261 RIMEDRKGQE NFQLIVITHD ERFAQMIGQR QHAEKYYRVA KDDMQHSIIE AQEIFD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
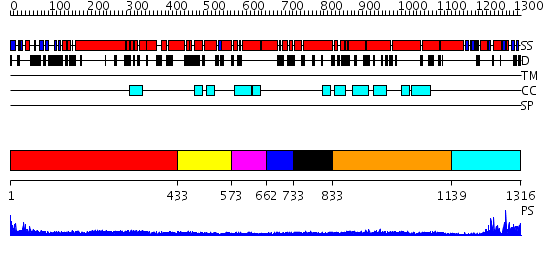
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..432] | 15.0 | Heavy meromyosin subfragment | |
| 2 | View Details | [433..572] | 9.69897 | Tropomyosin | |
| 3 | View Details | [573..661] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [662..732] | 16.102373 | No description for PF04423.5 was found. No confident structure predictions are available. | |
| 5 | View Details | [733..832] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [833..1138] | 2.12 | Tropomyosin | |
| 7 | View Details | [1139..1316] | 15.69897 | No description for 2ghiA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)