













| General Information: |
|
| Name(s) found: |
ISPD_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 302 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] plastid [NAS] |
| Biological Process: |
isopentenyl diphosphate biosynthetic process, mevalonate-independent pathway
[TAS]
|
| Molecular Function: |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMLQTNLGF ITSPTFLCPK LKVKLNSYLW FSYRSQVQKL DFSKRVNRSY KRDALLLSIK 60
61 CSSSTGFDNS NVVVKEKSVS VILLAGGQGK RMKMSMPKQY IPLLGQPIAL YSFFTFSRMP 120
121 EVKEIVVVCD PFFRDIFEEY EESIDVDLRF AIPGKERQDS VYSGLQEIDV NSELVCIHDS 180
181 ARPLVNTEDV EKVLKDGSAV GAAVLGVPAK ATIKEVNSDS LVVKTLDRKT LWEMQTPQVI 240
241 KPELLKKGFE LVKSEGLEVT DDVSIVEYLK HPVYVSQGSY TNIKVTTPDD LLLAERILSE 300
301 DS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
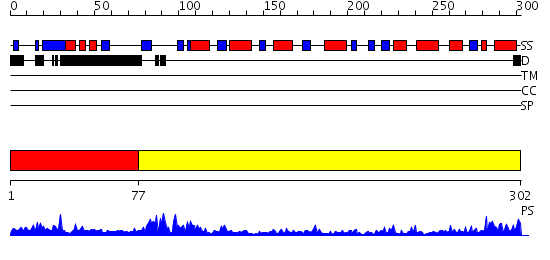
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | 6.0 | Crystal structure of UDP-N-acetylglucosamine pyrophosphorylase (Agx2) from Mus musculus at 2.50 A resolution | |
| 2 | View Details | [77..302] | 37.522879 | 2C-methyl-D-erythritol 4-phosphate cytidylyltransferase (IspD) from Arabidopsis thaliana |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)