













| General Information: |
|
| Name(s) found: |
GLYM1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 517 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
chloroplast [IDA] cytosolic ribosome [IDA] mitochondrial matrix [ISS] membrane [IDA] mitochondrion [IDA] apoplast [IDA] stromule [IDA] plasma membrane [IDA] |
| Biological Process: |
response to cold
[IEP]
oxygen and reactive oxygen species metabolic process [TAS] L-serine metabolic process [ISS] photorespiration [IMP] glycine decarboxylation via glycine cleavage system [IMP] response to cadmium ion [IEP] plant-type hypersensitive response [TAS] glycine metabolic process [ISS] |
| Molecular Function: |
glycine hydroxymethyltransferase activity
[ISS][TAS]
poly(U) RNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMAMALRRL SSSIDKPIRP LIRSTSCYMS SLPSEAVDEK ERSRVTWPKQ LNAPLEEVDP 60
61 EIADIIEHEK ARQWKGLELI PSENFTSVSV MQAVGSVMTN KYSEGYPGAR YYGGNEYIDM 120
121 AETLCQKRAL EAFRLDPEKW GVNVQPLSGS PANFHVYTAL LKPHERIMAL DLPHGGHLSH 180
181 GYQTDTKKIS AVSIFFETMP YRLDESTGYI DYDQMEKSAT LFRPKLIVAG ASAYARLYDY 240
241 ARIRKVCNKQ KAVMLADMAH ISGLVAANVI PSPFDYADVV TTTTHKSLRG PRGAMIFFRK 300
301 GVKEINKQGK EVLYDFEDKI NQAVFPGLQG GPHNHTITGL AVALKQATTS EYKAYQEQVL 360
361 SNSAKFAQTL MERGYELVSG GTDNHLVLVN LKPKGIDGSR VEKVLEAVHI ASNKNTVPGD 420
421 VSAMVPGGIR MGTPALTSRG FVEEDFAKVA EYFDKAVTIA LKVKSEAQGT KLKDFVSAME 480
481 SSSTIQSEIA KLRHEVEEFA KQFPTIGFEK ETMKYKN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
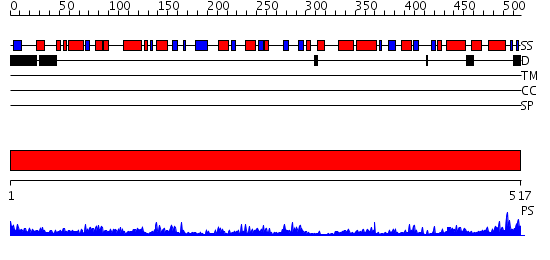
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..517] | 93.154902 | Human mitochondrial serine hydroxymethyltransferase 2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)