













| General Information: |
|
| Name(s) found: |
PHS1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 962 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] plastid [TAS] |
| Biological Process: |
response to temperature stimulus
[IEP]
response to water deprivation [IMP] |
| Molecular Function: |
phosphorylase activity
[IMP][ISS]
transferase activity, transferring glycosyl groups [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTMRISGVS TGAEVLIQCN SLSSLVSRRC DDGKWRTRMF PARNRDLRPS PTRRSFLSVK 60
61 SISSEPKAKV TDAVLDSEQE VFISSMNPFA PDAASVASSI KYHAEFTPLF SPEKFELPKA 120
121 FFATAQSVRD ALIMNWNATY EYYNRVNVKQ AYYLSMEFLQ GRALSNAVGN LGLNSAYGDA 180
181 LKRLGFDLES VASQEPDPAL GNGGLGRLAS CFLDSMATLN YPAWGYGLRY KYGLFKQRIT 240
241 KDGQEEAAED WLELSNPWEI VRNDVSYPIK FYGKVVFGSD GKKRWIGGED IVAVAYDVPI 300
301 PGYKTKTTIN LRLWSTKAPS EDFDLSSYNS GKHTEAAEAL FNAEKICFVL YPGDESTEGK 360
361 ALRLKQQYTL CSASLQDIVA RFETRSGGNV NWEEFPEKVA VQMNDTHPTL CIPELMRILM 420
421 DLKGLSWEDA WKITQRTVAY TNHTVLPEAL EKWSLELMEK LLPRHVEIIE KIDEELVRTI 480
481 VSEYGTADPD LLEEKLKAMR ILENVELPSA FADVIVKPVN KPVTAKDAQN GVKTEQEEEK 540
541 TAGEEEEDEV IPEPTVEPPK MVRMANLAVV GGHAVNGVAE IHSEIVKQDV FNDFVQLWPE 600
601 KFQNKTNGVT PRRWIRFCNP YLSDIITNWI GTEDWVLNTE KVAELRKFAD NEDLQSEWRA 660
661 AKKKNKLKVV SLIKERTGYT VSPDAMFDIQ IKRIHEYKRQ LLNILGIVYR YKKMKEMSAS 720
721 EREKAFVPRV CIFGGKAFAT YVQAKRIVKF ITDVASTINH DPEIGDLLKV IFVPDYNVSV 780
781 AELLIPASEL SQHISTAGME ASGTSNMKFS MNGCVLIGTL DGANVEIREE VGEENFFLFG 840
841 AKADQIVNLR KERAEGKFVP DPTFEEVKKF VGSGVFGSNS YDELIGSLEG NEGFGRADYF 900
901 LVGKDFPSYI ECQEKVDEAY RDQKRWTRMS IMNTAGSFKF SSDRTIHEYA KDIWNIKQVE 960
961 LP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
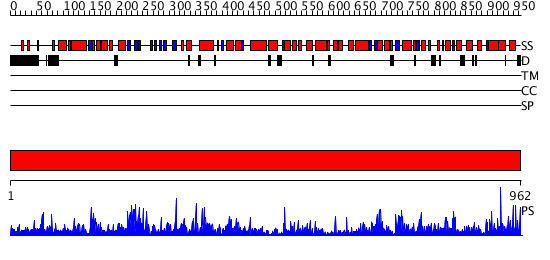
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..962] | 1000.0 | No description for 2zb2A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)