













| General Information: |
|
| Name(s) found: |
HDG11_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 722 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS]
|
| Biological Process: |
regulation of transcription, DNA-dependent
[ISS]
trichome branching [IGI][IMP] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFVVGVGGS GSGSGGDGGG SHHHDGSETD RKKKRYHRHT AQQIQRLESS FKECPHPDEK 60
61 QRNQLSRELG LAPRQIKFWF QNRRTQLKAQ HERADNSALK AENDKIRCEN IAIREALKHA 120
121 ICPNCGGPPV SEDPYFDEQK LRIENAHLRE ELERMSTIAS KYMGRPISQL STLHPMHISP 180
181 LDLSMTSLTG CGPFGHGPSL DFDLLPGSSM AVGPNNNLQS QPNLAISDMD KPIMTGIALT 240
241 AMEELLRLLQ TNEPLWTRTD GCRDILNLGS YENVFPRSSN RGKNQNFRVE ASRSSGIVFM 300
301 NAMALVDMFM DCVKWTELFP SIIAASKTLA VISSGMGGTH EGALHLLYEE MEVLSPLVAT 360
361 REFCELRYCQ QTEQGSWIVV NVSYDLPQFV SHSQSYRFPS GCLIQDMPNG YSKVTWVEHI 420
421 ETEEKELVHE LYREIIHRGI AFGADRWVTT LQRMCERFAS LSVPASSSRD LGGVILSPEG 480
481 KRSMMRLAQR MISNYCLSVS RSNNTRSTVV SELNEVGIRV TAHKSPEPNG TVLCAATTFW 540
541 LPNSPQNVFN FLKDERTRPQ WDVLSNGNAV QEVAHISNGS HPGNCISVLR GSNATHSNNM 600
601 LILQESSTDS SGAFVVYSPV DLAALNIAMS GEDPSYIPLL SSGFTISPDG NGSNSEQGGA 660
661 STSSGRASAS GSLITVGFQI MVSNLPTAKL NMESVETVNN LIGTTVHQIK TALSGPTAST 720
721 TA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
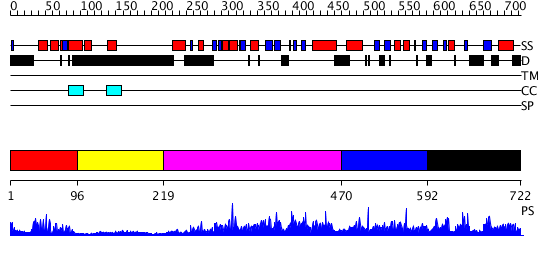
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..95] | 20.39794 | Paired protein | |
| 2 | View Details | [96..218] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [219..469] | 2.71 | No description for 2r55A was found. | |
| 4 | View Details | [470..591] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [592..722] | 3.062981 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.79 |
Source: Reynolds et al. (2008)