













| General Information: |
|
| Name(s) found: |
ANT_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 555 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC][IDA]
|
| Biological Process: |
regulation of transcription, DNA-dependent
[ISS][TAS]
gamete generation [IMP] regulation of cell proliferation [IMP] organ morphogenesis [IMP] |
| Molecular Function: |
DNA binding
[IDA][ISS][TAS]
transcription factor activity [ISS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKSFCDNDDN NHSNTTNLLG FSLSSNMMKM GGRGGREAIY SSSTSSAATS SSSVPPQLVV 60
61 GDNTSNFGVC YGSNPNGGIY SHMSVMPLRS DGSLCLMEAL NRSSHSNHHQ DSSPKVEDFF 120
121 GTHHNNTSHK EAMDLSLDSL FYNTTHEPNT TTNFQEFFSF PQTRNHEEET RNYGNDPSLT 180
181 HGGSFNVGVY GEFQQSLSLS MSPGSQSSCI TGSHHHQQNQ NQNHQSQNHQ QISEALVETS 240
241 VGFETTTMAA AKKKRGQEDV VVVGQKQIVH RKSIDTFGQR TSQYRGVTRH RWTGRYEAHL 300
301 WDNSFKKEGH SRKGRQVYLG GYDMEEKAAR AYDLAALKYW GPSTHTNFSA ENYQKEIEDM 360
361 KNMTRQEYVA HLRRKSSGFS RGASIYRGVT RHHQHGRWQA RIGRVAGNKD LYLGTFGTQE 420
421 EAAEAYDVAA IKFRGTNAVT NFDITRYDVD RIMSSNTLLS GELARRNNNS IVVRNTEDQT 480
481 ALNAVVEGGS NKEVSTPERL LSFPAIFALP QVNQKMFGSN MGGNMSPWTS NPNAELKTVA 540
541 LTLPQMPVFA AWADS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
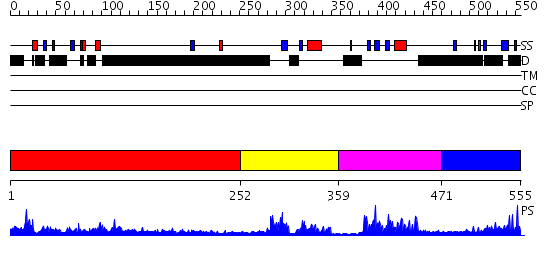
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..251] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [252..358] | 3.69 | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | |
| 3 | View Details | [359..470] | 23.522879 | GCC-box binding domain | |
| 4 | View Details | [471..555] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 |
|
||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)