













| General Information: |
|
| Name(s) found: |
PPIL2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 595 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[IEA]
|
| Biological Process: |
protein folding
[ISS]
protein ubiquitination [IEA] |
| Molecular Function: |
ubiquitin-protein ligase activity
[IEA]
peptidyl-prolyl cis-trans isomerase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKKQHSKDR MFITKTEWAT EWGGAKSKEN RTPFKSLPYY CCALTFLPFE DPVCTIDGSV 60
61 FEITTIVPYI RKFGKHPVTG APLKGEDLIP LIFHKNSEGE YHCPVLNKVF TEFTHIVAVK 120
121 TTGNVFCYEA IKELNIKTKN WKELLTEEPF TRADLITIQN PNAVDGKVTV EFDHVKNGLK 180
181 IDDEELKKMN SDPAYNINVS GDIKHMLADL GTDKAKEIAL HGGGGNKARN ERAAAIAAIL 240
241 ESRSKIKEVS KAEQPKQTYS VVDAASASVF GRSADAAKAG SSDKTAARIA MHMAGDRTPV 300
301 NSKMVKSRYS SGAASRSFTS SAFTPVTKND FELIKVEKNP KKKGYVQFQT THGDLNIELH 360
361 CDIAPRACEN FITLCERGYY NGVAFHRSIR NFMIQGGDPT GTGKGGESIW GKPFKDEPNS 420
421 KLLHSGRGVV SMANSGPHTN GSQFFVLYKS ATHLNYKHTV FGGVVGGLAT LAAMENVPVD 480
481 ESDRPLEEIK IIEASVFVNP YTELDEEEEK EKAEKEKNED KDIEKIGSWY SNPGSGTTEA 540
541 GAGGGGVGKY LKAMSSTATK DTKGSLDSDI STIGVSKKRK TTASASTGFK DFSSW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
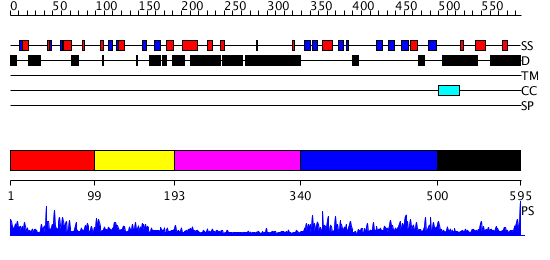
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..98] | 15.69897 | No description for 2f42A was found. | |
| 2 | View Details | [99..192] | 2.19 | Not-4 N-terminal RING finger domain | |
| 3 | View Details | [193..339] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [340..499] | 55.69897 | Crystal Structure of the cyclophiln_RING domain of human peptidylprolyl isomerase (cyclophilin)-like 2 isoform b | |
| 5 | View Details | [500..595] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)