













| General Information: |
|
| Name(s) found: |
DEGP9_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 592 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
nucleolus [IDA] |
| Biological Process: |
proteolysis
[IEA][ISS]
|
| Molecular Function: |
protein binding
[IEA]
catalytic activity [IEA] serine-type endopeptidase activity [IEA] serine-type peptidase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKNSEKRGRK HKRQDASSAE NAGGEVKEAS ANEASLPQSP EPVSASEANP SPSRRSRGRG 60
61 KKRRLNNESE AGNQRTSSPE RSRSRLHHSD TKNGDCSNGM IVSTTTESIP AAPSWETVVK 120
121 VVPSMDAVVK VFCVHTEPNF SLPWQRKRQY SSGSSGFIIG GRRVLTNAHS VEHHTQVKLK 180
181 KRGSDTKYLA TVLAIGTECD IALLTVTDDE FWEGVSPVEF GDLPALQDAV TVVGYPIGGD 240
241 TISVTSGVVS RMEILSYVHG STELLGLQID AAINSGNSGG PAFNDKGKCV GIAFQSLKHE 300
301 DAENIGYVIP TPVIVHFIQD YEKHDKYTGF PVLGIEWQKM ENPDLRKSMG MESHQKGVRI 360
361 RRIEPTAPES QVLKPSDIIL SFDGVNIAND GTVPFRHGER IGFSYLISQK YTGDSALVKV 420
421 LRNKEILEFN IKLAIHKRLI PAHISGKPPS YFIVAGFVFT TVSVPYLRSE YGKEYEFDAP 480
481 VKLLEKHLHA MAQSVDEQLV VVSQVLVSDI NIGYEEIVNT QVVAFNGKPV KNLKGLAGMV 540
541 ENCEDEYMKF NLDYDQIVVL DTKTAKEATL DILTTHCIPS AMSDDLKTEE RN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
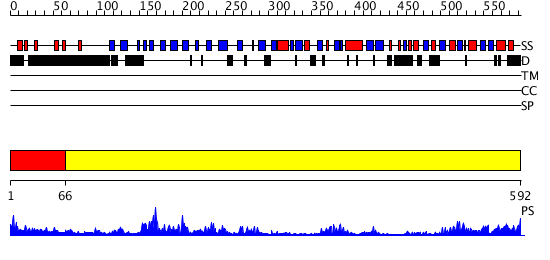
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..65] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [66..592] | 63.39794 | Protease Do (DegP, HtrA), C-terminal domains; Protease Do (DegP, HtrA), catalytic domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)