













| General Information: |
|
| Name(s) found: |
ISOA1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 783 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
chloroplast isoamylase complex [IDA] |
| Biological Process: |
carbohydrate metabolic process
[ISS]
amylopectin biosynthetic process [IMP] |
| Molecular Function: |
alpha-amylase activity
[ISS]
isoamylase activity [IDA][IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDAIKCSSSF LHHTKLNTLF SNHTFPKISA PNFKPLFRPI SISAKDRRSN EAENIAVVEK 60
61 PLKSDRFFIS DGLPSPFGPT VRDDGVNFSV YSTNSVSATI CLISLSDLRQ NKVTEEIQLD 120
121 PSRNRTGHVW HVFLRGDFKD MLYGYRFDGK FSPEEGHYYD SSNILLDPYA KAIISRDEFG 180
181 VLGPDDNCWP QMACMVPTRE EEFDWEGDMH LKLPQKDLVI YEMHVRGFTR HESSKIEFPG 240
241 TYQGVAEKLD HLKELGINCI ELMPCHEFNE LEYYSYNTIL GDHRVNFWGY STIGFFSPMI 300
301 RYASASSNNF AGRAINEFKI LVKEAHKRGI EVIMDVVLNH TAEGNEKGPI FSFRGVDNSV 360
361 YYMLAPKGEF YNYSGCGNTF NCNHPVVRQF ILDCLRYWVT EMHVDGFRFD LGSIMSRSSS 420
421 LWDAANVYGA DVEGDLLTTG TPISCPPVID MISNDPILRG VKLIAEAWDA GGLYQVGMFP 480
481 HWGIWSEWNG KFRDVVRQFI KGTDGFSGAF AECLCGSPNL YQGGRKPWHS INFICAHDGF 540
541 TLADLVTYNN KNNLANGEEN NDGENHNYSW NCGEEGDFAS ISVKRLRKRQ MRNFFVSLMV 600
601 SQGVPMIYMG DEYGHTKGGN NNTYCHDNYM NYFRWDKKEE AHSDFFRFCR ILIKFRDECE 660
661 SLGLNDFPTA KRLQWHGLAP EIPNWSETSR FVAFSLVDSV KKEIYVAFNT SHLATLVSLP 720
721 NRPGYRWEPF VDTSKPSPYD CITPDLPERE TAMKQYRHFL DANVYPMLSY SSIILLLSPI 780
781 KDP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
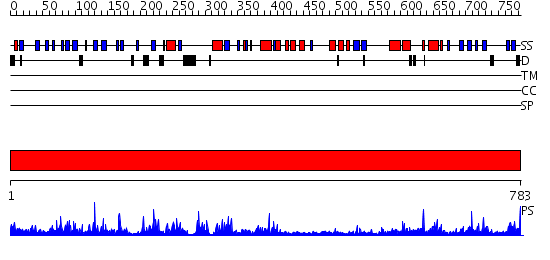
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..783] | 93.0 | STRUCTURE OF PSEUDOMONAS ISOAMYLASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)