













| General Information: |
|
| Name(s) found: |
HIBC1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 378 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to cold
[IMP]
fatty acid beta-oxidation [TAS] valine catabolic process [IDA] response to auxin stimulus [IMP] |
| Molecular Function: |
3-hydroxyisobutyryl-CoA hydrolase activity
[IGI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVEMASQSQ VLVEEKSSVR ILTLNRPKQL NALSFHMISR LLQLFLAFEE DPSVKLVILK 60
61 GHGRAFCAGG DVAAVVRDIN QGNWRLGANY FSSEYMLNYV MATYSKAQVS ILNGIVMGGG 120
121 AGVSVHGRFR IATENTVFAM PETALGLFPD VGASYFLSRL PGFFGEYVGL TGARLDGAEM 180
181 LACGLATHFV PSTRLTALEA DLCRINSNDP TFASTILDAY TQHPRLKQQS AYRRLDVIDR 240
241 CFSRRTVEEI ISALEREATQ EADGWISATI QALKKGSPAS LKISLRSIRE GRLQGVGQCL 300
301 IREYRMVCHV MKGEISKDFV EGCRAILVDK DKNPKWEPRR LEDMKDSMVE QYFERVERED 360
361 DLKLPPRNNL PALGIAKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
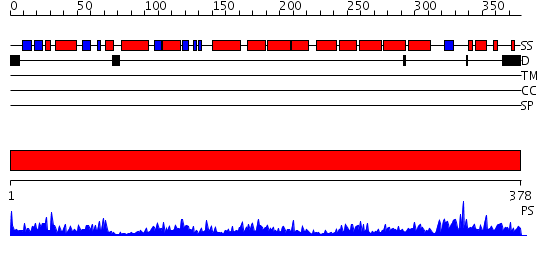
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..378] | 48.0 | fatty acid beta-oxidation multienzyme complex from Pseudomonas fragi, form I (native2) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)