













| General Information: |
|
| Name(s) found: |
GLYC4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 471 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IDA]
cytosol [ISS] plasma membrane [IDA] |
| Biological Process: |
L-serine metabolic process
[ISS]
response to cadmium ion [IEP] glycine metabolic process [ISS] |
| Molecular Function: |
pyridoxal phosphate binding
[IEA]
catalytic activity [IEA] glycine hydroxymethyltransferase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEPVSSWGNT SLVSVDPEIH DLIEKEKRRQ CRGIELIASE NFTSFAVIEA LGSALTNKYS 60
61 EGIPGNRYYG GNEFIDEIEN LCRSRALEAF HCDPAAWGVN VQPYSGSPAN FAAYTALLQP 120
121 HDRIMGLDLP SGGHLTHGYY TSGGKKISAT SIYFESLPYK VNFTTGYIDY DKLEEKALDF 180
181 RPKLLICGGS AYPRDWDYAR FRAIADKVGA LLLCDMAHIS GLVAAQEAAN PFEYCDVVTT 240
241 TTHKSLRGPR AGMIFYRKGP KPPKKGQPEG AVYDFEDKIN FAVFPALQGG PHNHQIGALA 300
301 VALKQANTPG FKVYAKQVKA NAVALGNYLM SKGYQIVTNG TENHLVLWDL RPLGLTGNKV 360
361 EKLCDLCSIT LNKNAVFGDS SALAPGGVRI GAPAMTSRGL VEKDFEQIGE FLSRAVTLTL 420
421 DIQKTYGKLL KDFNKGLVNN KDLDQLKADV EKFSASYEMP GFLMSEMKYK D |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
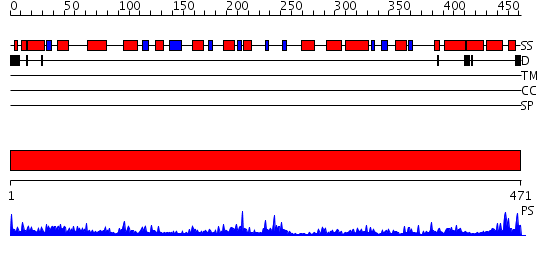
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..471] | 105.0 | Human mitochondrial serine hydroxymethyltransferase 2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)