













| General Information: |
|
| Name(s) found: |
gi|25372680
[NCBI NR]
gi|15220007 [NCBI NR] gi|12324592 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 882 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
[IEA]
|
| Molecular Function: |
nucleic acid binding
[IEA]
DNA binding [IEA] ATP binding [IEA] hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides [IEA] ATP-dependent helicase activity [IEA] ATP-dependent DNA helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEIREFPAF PYKPYSIQID FMNALYQFLD KGGVSMLESP TGTGKSLSII CSALQWLTDR 60
61 KEKRNSGLTE ESHKVDGNDD EDEPDWMRNF TLNEADGANG DKSIKNTKSP FSLRKHAKEV 120
121 FTEGEKADEL NDQEFLLDEY ESQDESSPGG GNSKRKPARG FDSSSSEDEE DENDCSDDEQ 180
181 GGFKVFFCSR THSQLSQFVK ELRKTVFAKK LKVVCLGSRK NLCINEDVLK LGNVTRINER 240
241 CLDLQKKKIS QVSKKKNLGA NVRIVRTKAS CRCPMLRKHK LQREFKAESF QQEAMDIEDL 300
301 VQLGREMRTC PYYGSRRMAP AADLVILPYQ SLLSKSSRES LGLSLKNSVV IIDEAHNLAD 360
361 TLLSMHDAKL TVSQLEDIHC SIESYLGRFQ NLLGAGNRRY IQIMLILTRA FLKPLASDRN 420
421 LSSVNVGLDT ENPSKSKPCG ACSMAIYDFL FSLNIDNINL VKLLAYIKES NIIHKVSGYG 480
481 ERVAAMLQKD PVAHEEMSKL TSFRAFSDML VALTNNNGDG RIIISRTSSS TSGQHGGYIK 540
541 YVMLTGAKLF SEVVDEAHAV ILAGGTLQPI EETRERLFPW LPSNQLQFFS CSHIVPPESI 600
601 MPIAVSHGPS GQSFDFSHSS RSSIGMIQEL GLLMSNLVAV VPEGIIVFFS SFEYETQVHT 660
661 AWSNSGILRR IVKKKRVFRE PRKNTEVEAV LRDYKEAIES ERGAIMLAVV GGKVSEGINF 720
721 SDSMCRCVVM VGLPYPSPSD IELLERIKHI EGRADSDSIK PSVTLVDDSY YSGDVQAGFG 780
781 VLKSCKRRGK EYYENLCMKA VNQSIGRAIR HEKDYASILL VDARYSNDPS KRTSHSHPSN 840
841 KLPKWIKDRL IYSTKGYGDV HRLLHQFFKH KNVENSSETN QH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
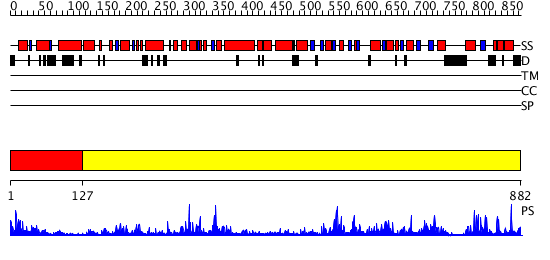
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..126] | 5.294997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [127..882] | 68.69897 | No description for 2vsfA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)