













| General Information: |
|
| Name(s) found: |
DEXHD_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 2172 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleotide binding
[IEA]
nucleic acid binding [IEA] ATP binding [IEA] ATP-dependent helicase activity [IEA][ISS] nucleoside-triphosphatase activity [IEA] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTNLGGGGAE EQARLKQYGY KVNSSLVLNS DERRRDTHES SGEPESLRGR IDPKSFGDRV 60
61 VRGRPHELDE RLNKSKKKKE RCDDLVSARE SKRVRLREVS VLNDTEDGVY QPKTKETRVA 120
121 FEIMLGLIQQ QLGGQPLDIV CGAADEILAV LKNESVKNHE KKVEIEKLLN VITDQVFSQF 180
181 VSIGKLITDY EEGGDSLSGK ASEDGGLDYD IGVALECEED DDESDLDMVQ DEKDEEDEDV 240
241 VELNKTGVVQ VGVAINGEDA RQAKEDTSLN VLDIDAYWLQ RKISQEYEQK IDAQECQELA 300
301 EELLKILAEG NDRDVEIKLL EHLQFEKFSL VKFLLQNRLK VVWCTRLARG RDQEERNQIE 360
361 EEMLGLGSEL AAIVKELHAK RATAKEREEK REKDIKEEAQ HLMDDDSGVD GDRGMRDVDD 420
421 LDLENGWLKG QRQVMDLESL AFNQGGFTRE NNKCELPDRS FRIRGKEFDE VHVPWVSKKF 480
481 DSNEKLVKIS DLPEWAQPAF RGMQQLNRVQ SKVYGTALFK ADNILLCAPT GAGKTNVAVL 540
541 TILHQLGLNM NPGGTFNHGN YKIVYVAPMK ALVAEVVDSL SQRLKDFGVT VKELSGDQSL 600
601 TGQEIKETQI IVTTPEKWDI ITRKSGDRTY TQLVRLLIID EIHLLDDNRG PVLESIVART 660
661 LRQIESTKEH IRLVGLSATL PNCDDVASFL RVDLKNGLFI FDRSYRPVPL GQQYIGINVK 720
721 KPLRRFQLMN DICYQKVVAV AGKHQVLIFV HSRKETAKTA RAIRDTAMAN DTLSRFLKED 780
781 SQSREILKCL AGLLKNNDLK ELLPYGFAIH HAGLTRTDRE IVENQFRWGN LQVLISTATL 840
841 AWGVNLPAHT VIIKGTQVYN PERGEWMELS PLDVMQMIGR AGRPQYDQQG EGIIITGYSK 900
901 LQYYLRLMNE QLPIESQFIS KLADQLNAEI VLGTIQNARE ACHWLGYTYL YVRMVRNPTL 960
961 YGVSPDALAK DLLLEERRAD LIHSAATILD KNNLIKYDRK SGHFQVTDLG RIASYYYISH1020
1021 GTIAAYNENL KPTMNDIELC RLFSLSEEFK YVTVRQDEKM ELAKLLDRVP IPVKETLEDP1080
1081 SAKINVLLQV YISKLKLEGL SLTSDMVYIT QSAGRLLRAI FEIVLKRGWA QLSQKALNLS1140
1141 KMVGKRMWSV QTPLWQFPGI PKEILMKLEK NDLVWERYYD LSSQELGELI CNPKMGRPLH1200
1201 KYIHQFPKLK LAAHVQPISR SVLQVELTVT PDFHWDDKAN KYVEPFWIIV EDNDGEKILH1260
1261 HEYFLFKKRV IDEDHTLNFT VPISEPIPPQ YFIRVVSDKW LDSPTVLPVS FRHLILPEKY1320
1321 PPPTELLDLQ PLPVMALRNP SYETLYQDFK HFNPVQTQVF TVLYNTSDNV VVAAPTGSGK1380
1381 TICAEFAILR NHLEGPDSAM RVVYIAPLEA IAKEQFRDWE KKFGKGLGLR VVELTGETLL1440
1441 DLKLLEKGQI IISTPEKWDA LSRRWKQRKY IQQVSLFIVD ELHLIGGQGG QVLEVIVSRM1500
1501 RYISSQVGNK IRIVALSTSL ANAKDLGEWI GASSCGVFNF PPNVRPVPLE IHIHGVDILS1560
1561 FEARMQAMTK PTYTAIVQHA KNKKPAIVFV PTRKHVRLTA VDLIAYSHMD NMKSPDFLLG1620
1621 NLEELEPFLI QICEETLKET LRHGIGYLHE GLSNLDQEIV TQLFEAGRIQ VCVMSSSLCW1680
1681 GTPLKAHLVV VMGTHFYDGR ENSHSDYPIS NLLQMMGRGS RPLLDDAGKC VIFCHAPRKE1740
1741 YYKKFLYEAL PVESHLQHFL HDNFNAEVVA RVIENKQDAV DYLTWSFMYR RLPQNPNYYN1800
1801 LLGVSHRHLS DHLSELVENT LSDLEVSKCI EIDNELDLSP LNLGMIASYY YINYTTIERF1860
1861 SSLLASKTKM KGLLEILTSA SEYDLIPIRP GEEDAVRRLI NHQRFSFQNP RCTDPRVKTS1920
1921 ALLQAHFSRQ KISGNLVMDQ CEVLLSATRL LQAMVDVISS NGCLNLALLA MEVSQMVTQG1980
1981 MWDRDSMLLQ LPHFTKDLAK RCHENPGNNI ETIFDLVEME DDKRQELLQM SDAQLLDIAR2040
2041 FCNRFPNIDL TYEIVGSNEV SPGKDITLQV LLERDMEGRT EVGPVDAPRY PKTKEEGWWL2100
2101 VVGEAKTNQL MAIKRISLQR KAQVKLEFAV PTETGEKSYT LYFMCDSYLG CDQEYSFTVD2160
2161 VKDSDAADHM EE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
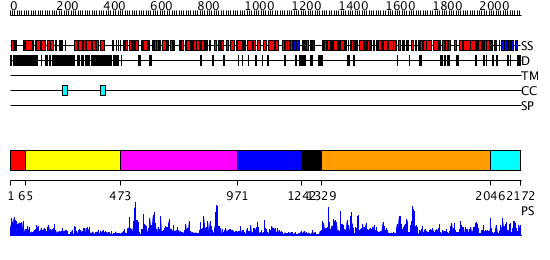
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..64] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [65..472] | 7.69897 | Heavy meromyosin subfragment | |
| 3 | View Details | [473..970] | 85.522879 | No description for 2i4iA was found. | |
| 4 | View Details | [971..1241] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1242..1328] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1329..2045] | 86.045757 | No description for 2va8A was found. | |
| 7 | View Details | [2046..2172] | 88.39794 | No description for 2q0zX was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)