













| General Information: |
|
| Name(s) found: |
FIMB3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 714 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuole
[IDA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
actin binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGFVGVIVS DPWLQSQLTQ VELRSLNSKF VALKNQSGKV TLEDLPSVLV KVKSLSSSFK 60
61 EKEIKEILGG LGSDYESDDD LDFESFLKVY LNLRDKAADK AGGGLKHSSS FLKAGTTTLH 120
121 TINQSEKGSF VLHINRYLGD DPFLKQFLPL DPDSNDLYEL VKDGVLLCKL INIAVPGTID 180
181 ERAINTKRVL NPWERNENHT LCLNSAKAVG CSVVNIGTQD LAEGRPHLVL GLISQLIKIQ 240
241 LLADLSLKKM PQLVELVEDN EDIEEFLRLP PEKVLLKWMN FHLKKGGYKK TVGNFSSDLK 300
301 DAQAYAYLLN VLAPEHCDPA TLNAEDDLER ANMVLEHAER MNCKRYLTAE EIVEGSSYLN 360
361 LAFVAQIFHE RNGLSTDGRF SFAEMMTEDL QTCRDERCYR LWINSLGIES YVNNVFEDVR 420
421 NGWILLEVVD KVYPGSVNWK QASKPPIKMP FRKVENCNQV VKIGKEMRFS LVNVAGNDIV 480
481 QGNKKLILGF LWQLMRTHML QLLKSLRSRT RGKDMTDSEI ISWANRKVRI MGRKSQIESF 540
541 KDKSLSSGLF FLDLLWAVEP RVVNWNLVTK GESDDEKRLN ATYIVSVARK LGCSVFLLPE 600
601 DIVEVNQKMI LILTASIMYW SLQQQSSSSE SSSSSSDSSS THSTTTTCTS TCTSTDASPA 660
661 PSVTGEDEVS SLNGEVSSLT IEEDNEVSSL TIEEDNDADI LSDITSISEE AANE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
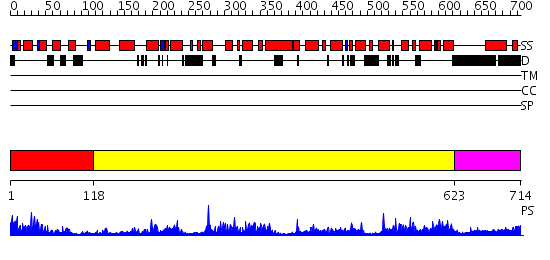
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | 23.69897 | Structure of calmodulin bound to a calcineurin peptide: a new way of making an old binding mode | |
| 2 | View Details | [118..622] | 115.0 | Crystal structure of the actin-crosslinking core of Arabidopsis fimbrin | |
| 3 | View Details | [623..714] | 66.154902 | Cryo-EM Structure of Chicken Gizzard Smooth Muscle alpha-Actinin |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)