













| General Information: |
|
| Name(s) found: |
LHP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 445 amino acids |
Gene Ontology: |
|
| Cellular Component: |
euchromatin
[IDA]
nuclear heterochromatin [IDA] |
| Biological Process: |
photoperiodism, flowering
[IMP]
[NOT]root development [IMP] negative regulation of gene expression, epigenetic [IMP] multidimensional cell growth [IMP] shoot morphogenesis [IMP] chromatin silencing [IEP][IMP] chromatin organization [IMP] photoperiodism [IMP] vernalization response [IMP] negative regulation of flower development [IMP] |
| Molecular Function: |
DNA binding
[IDA]
methylated histone residue binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKGASGAVKK KPQVLNEAGE AETAVETVGE SRKISGDGGF GSDDGGGGGG GGSGESILRE 60
61 IGDDRPTEDG DEEEEEDEDE DDGGDEEDEE GEGEGGQEER PKLDEGFYEI EAIRRKRVRK 120
121 GKVQYLIKWR GWPETANTWE PLENLQSIAD VIDAFEGSLK PGKPGRKRKR KYAGPHSQMK 180
181 KKQRLTSTSH DATEKSDSST SLNNSSLPDI PDPLDLSGSS LLNRDVEAKN AYVSNQVEAN 240
241 SGSVGMARQV RLIDNEKEYD PTLNELRGPV NNSNGAGCSQ GGGIGSEGDN VRPNGLLKVY 300
301 PKELDKNSRF IGAKRRKSGS VKRFKQDGST SNNHTAPTDQ NLTPDLTTLD SFGRIARMGN 360
361 EYPGVMENCN LSQKTKIEEL DITKILKPMS FTASVSDNVQ EVLVTFLALR SDGKEALVDN 420
421 RFLKAHNPHL LIEFYEQHLK YNRTP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
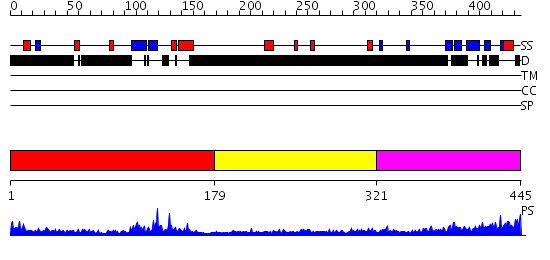
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..178] | 9.30103 | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and dimethylarginine 2 | |
| 2 | View Details | [179..320] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [321..445] | 1.27 | Heterochromatin protein 1, HP1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)