













| General Information: |
|
| Name(s) found: |
BGAL5_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 732 amino acids |
Gene Ontology: |
|
| Cellular Component: |
beta-galactosidase complex
[IEA]
endomembrane system [IEA] |
| Biological Process: |
carbohydrate metabolic process
[IEA]
|
| Molecular Function: |
catalytic activity
[IEA]
cation binding [IEA] beta-galactosidase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTTILVLSK ILTFLLTTML IGSSVIQCSS VTYDKKAIVI NGHRRILLSG SIHYPRSTPE 60
61 MWEDLIKKAK DGGLDVIDTY VFWNGHEPSP GTYNFEGRYD LVRFIKTIQE VGLYVHLRIG 120
121 PYVCAEWNFG GFPVWLKYVD GISFRTDNGP FKSAMQGFTE KIVQMMKEHR FFASQGGPII 180
181 LSQIENEFEP DLKGLGPAGH SYVNWAAKMA VGLNTGVPWV MCKEDDAPDP IINTCNGFYC 240
241 DYFTPNKPYK PTMWTEAWSG WFTEFGGTVP KRPVEDLAFG VARFIQKGGS YINYYMYHGG 300
301 TNFGRTAGGP FITTSYDYDA PIDEYGLVQE PKYSHLKQLH QAIKQCEAAL VSSDPHVTKL 360
361 GNYEEAHVFT AGKGSCVAFL TNYHMNAPAK VVFNNRHYTL PAWSISILPD CRNVVFNTAT 420
421 VAAKTSHVQM VPSGSILYSV ARYDEDIATY GNRGTITARG LLEQVNVTRD TTDYLWYTTS 480
481 VDIKASESFL RGGKWPTLTV DSAGHAVHVF VNGHFYGSAF GTRENRKFSF SSQVNLRGGA 540
541 NKIALLSVAV GLPNVGPHFE TWATGIVGSV VLHGLDEGNK DLSWQKWTYQ AGLRGESMNL 600
601 VSPTEDSSVD WIKGSLAKQN KQPLTWYKAY FDAPRGNEPL ALDLKSMGKG QAWINGQSIG 660
661 RYWMAFAKGD CGSCNYAGTY RQNKCQSGCG EPTQRWYHVP RSWLKPKGNL LVLFEELGGD 720
721 ISKVSVVKRS VN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
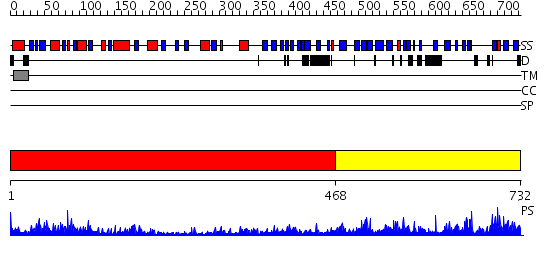
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..467] | 52.0 | Native structure of beta-galactosidase from Penicillium sp. | |
| 2 | View Details | [468..732] | 1.62 | beta-Glucuronidase; beta-Glucuronidase, domain 3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|