













| General Information: |
|
| Name(s) found: |
gi|7438357
[NCBI NR]
gi|41619196 [NCBI NR] gi|3298534 [NCBI NR] gi|20197429 [NCBI NR] gi|15225687 [NCBI NR] gi|111074230 [NCBI NR] gi|110738559 [NCBI NR] |
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 490 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS]
|
| Biological Process: |
regulation of transcription, DNA-dependent
[ISS]
gibberellic acid mediated signaling pathway [TAS] positive regulation of abscisic acid mediated signaling pathway [IMP] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS][TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGGGETTAT ATMEGRGLKK GPWTTTEDAI LTEYVRKHGE GNWNAVQKNS GLLRCGKSCR 60
61 LRWANHLRPN LKKGSFTPDE EKIIIDLHAK LGNKWARMAS QLPGRTDNEI KNYWNTRMKR 120
121 RQRAGLPLYP HEIQHQGIDI DDEFEFDLTS FQFQNQDLDH NHQNMIQYTN SSNTSSSSSS 180
181 FSSSSSQPSK RLRPDPLVST NPGLNPIPDS SMDFQMFSLY NNSLENDNNQ FGFSVPLSSS 240
241 SSSNEVCNPN HILEYISENS DTRNTNKKDI DAMSYSSLLM GDLEIRSSSF PLGLDNSVLE 300
301 LPSNQRPTHS FSSSPIIDNG VHLEPPSGNS GLLDALLEES QALSRGGLFK DVRVSSSDLC 360
361 EVQDKRVKMD FENLLIDHLN SSNHSSLGAN PNIHNKYNEP TMVKVTVDDD DELLTSLLNN 420
421 FPSTTTPLPD WYRVTEMQNE ASYLAPPSGI LMGNHQGNGR VEPPTVPPSS SVDPMASLGS 480
481 CYWSNMPSIC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
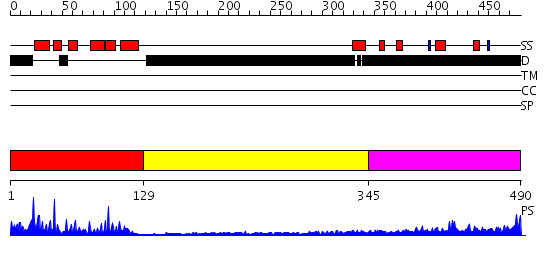
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] | 41.69897 | v-Myb | |
| 2 | View Details | [129..344] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [345..490] | 6.148993 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)