













| General Information: |
|
| Name(s) found: |
CDT1A_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 571 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
chloroplast [IDA] |
| Biological Process: |
DNA replication
[IMP]
chloroplast organization [IMP] |
| Molecular Function: |
protein binding
[IPI]
cyclin-dependent protein kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTPGSSRSI PFKSKKRLVM DSPSSKSQTG NPNPSSVALP TPEKPLENML SRSRNRSVAL 60
61 SVKEIRQAAG SRRRSEDPVA SSAKSRLFFD SSSSSPSKRK SSNKNAEKEK LPEKYENLGK 120
121 FFEALDNSML LSKLRGSKPT FSNISKQIEH LTERRFCYSH LAQIKHILPE AIEIKRVLIH 180
181 DETTCCMKPD LHVTLNADAV EYNDKSKSES KKIALRKVFR ARLADFVKAH PQGDEVPEEP 240
241 LPEPFNRRKP VENSNVEVKR VSSLMEEMAS IPASKLFSSP ITSTPVKTTS SLAKPTSSQI 300
301 NIAPTPTKPT STPAKQTLSE INILPTPVKP VSTLAKFPST PAIIDSTPVI TATPPEFAST 360
361 PARLMSTSLA ARPLKRSNGH TNPDDISADP PTKLVRRSLS LNFDSYPEDE RTMDFTDDIP 420
421 IDQVPEEDVS SDDEILSILP DKLRHAIKEQ ERKAIEDQNP AISLAKRRRK MIACLPKLFN 480
481 VIHYLIQSIR RWVITKEELV HKIIAGHSDI TDRKEVEEQL ILLQEIVPEW MSEKKSSSGD 540
541 VLVCINKLAS PLTIRSRLEE ENKQEMAPLL S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
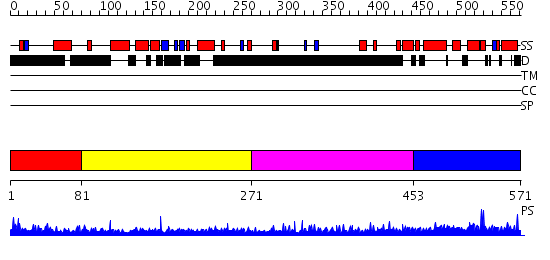
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..270] | 35.39794 | Strucure of Geminin-Cdt1 complex | |
| 3 | View Details | [271..452] | 6.169997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [453..571] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)