













| General Information: |
|
| Name(s) found: |
AGT23_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 481 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IEA]
|
| Biological Process: |
cellular response to nitrogen levels
[IEP]
|
| Molecular Function: |
pyridoxal phosphate binding
[IEA]
catalytic activity [IEA] transaminase activity [IEA] alanine-glyoxylate transaminase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRKLTAVNSL LKRNNYLLPR HGSSQTAAQR TSSVRETETE TKLPKMPPFN YSPPPYDGPS 60
61 TAEIIAKRRE FLSPALFHFY NTPLNIVEAK MQYVFDENGR RYLDAFGGIA TVSCGHCHPE 120
121 VVNSVVKQLK LINHSTILYL NHTISDFAEA LVSTLPGDLK VVFFTNSGTE ANELAMMMAR 180
181 LYTGCNDIVS LRNSYHGNAA ATMGATAQSN WKFNVVQSGV HHAINPDPYR GIFGSDGEKY 240
241 ASDVHDLIQF GTSGQVAGFI GESIQGVGGI VELAPGYLPA AYDIVRKAGG VCIADEVQSG 300
301 FARTGTHFWG FQSHGVIPDI VTMAKGIGNG IPLGAVVTTP EIAGVLSRRS YFNTFGGNPM 360
361 CTAAGHAVLR VLHEEKLQEN ANLVGSHLKR RLTLLKNKYE LIGDVRGRGL MLGVEFVKDR 420
421 DLKTPAKAET LHLMDQMKEM GVLVGKGGFY GNVFRITPPL CFTLSDADFL VDVMDHAMSK 480
481 M |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
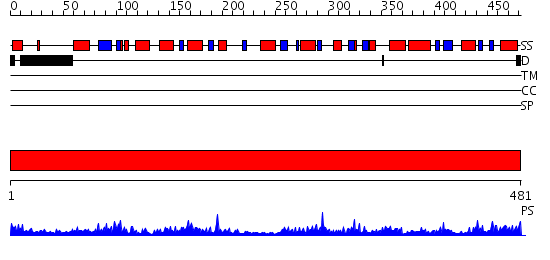
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..481] | 97.30103 | Structure of E. coli gamma-aminobutyrate aminotransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)