













| General Information: |
|
| Name(s) found: |
APC1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1678 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[TAS]
|
| Biological Process: |
ubiquitin-dependent protein catabolic process
[TAS]
|
| Molecular Function: |
ubiquitin-protein ligase activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPPGVRQLTV LGKFKPFGLI AEATDGKSPD DSYQYFLFDP ELTGERDDAD GNDANFSRQR 60
61 EHELFIRDNC NVRKGYALVE ANFIGYWYNA CWSNLGRGTE AFLCVLQIAC LTIYNTSGEV 120
121 VSVPLMRTVK SIWPLPCGLL LEQAGEVNPP SHVPFSPVSP ILGSREMLRQ RKEVGNSSPQ 180
181 NFHSPVAHDL ISKRDMPCMS SHLILRDPLE EPGPTYVEER GKLTIMKDYD ERTIWTSDRL 240
241 PLMTSYNKGK MQHSVWAAEF IESNLEASAS CSSGIVPDAV LSKRVSFRRI WQAKGAKKAA 300
301 SKVFLATDNS VPVICFLILE QKKLLSVGLQ TVEINNEILF DVKPDISWSV SAIAAAPVVV 360
361 TRSQVKIGLL PHLDIIVLSP ENDLFLYSGK QCLCRYVLPS WLGESIGSGD GESAKTDSGF 420
421 RNLKITGLSD AVLGSINLSV NHSQIFRCAL TGKPSSSLAN DCIAAIAEGL RSDLYSLFLS 480
481 LLWGDGHSDL QGSSIHFEWE ALCNIFLEIC QKPTVVHRKQ PKTASESSWE FLLISKFHKT 540
541 YSRFHNGITS INRLDLEGIV PFDSKICSEE TLGSSCELMV QSLDCLHAVY ESLKMDNLRK 600
601 QDLHHLAVLL CNIAKFLDEK CYLDYYIRDF PRLSTTIGAC TTLSSSRKPP NLFRWLENCL 660
661 RRGCLSTNFD DLPDLIRRDG CSIVSWARKV VSFYSVLFGD KPEGRTLSSG VPCNIAPGSY 720
721 SCNEELTILA MAGERFGLHQ LDLLPSGVSL PLRHALDSCR ESPPADWPAI AYVLLGREDM 780
781 ALSVFRNFSS SKEFEMQSNT SLISMSIPYM LHLHPVIVPS SESIGLENTK IEDTNSVDGS 840
841 VIDGMEHIFN SYTQLRYGRD LRLNEVRRLL CSARPVVVQT AANPTISDQE QQQAFTVPKL 900
901 VLAGRLPSQQ NAIVNLDPNI RNIQELKTWP EFHNAVAAGL RLAPLQGKVS RTWIRYNKPG 960
961 EPNAVHAGLL FGLGLQGYLH VLNLSDIYQY FTQDHESTTV GLMLGLAASY RGTMQPDIAK1020
1021 ALFFHVPARY QASYTEFEIP TLLQSAALVS VGMLFEGSAH QQTMQLLLGE IGRRSAGDNV1080
1081 LEREGYAVSA GFSLGLVALG RGGDALGSMD SLVNRLLLYL GAKEERSILV PSLEDHRSAA1140
1141 QITDGSTSNV DITAPGAIIA LTLMYLKTES EVIFSKLSIP QTHYDLECVR PDFIMLRVIA1200
1201 RNLIMWSRIC PTCDWIQSQV PEVVKNGISQ LRDDMDNMYE VDVEALVQAY VNIVAGACIS1260
1261 LGLRFAGTRD GNARDLLNSY ALYLLNEIKP LSATPGNAFP RGISKFVDRG TLEMCLYLII1320
1321 ISLSVVMAGS GDLQVFRLLR FLRSRNSADG HANYGTQMAV SLATGFLFLG GGMRTFSTNN1380
1381 GSLAMLLITL YPRLPSGPND NRCHLQAFRH LYVLATEARW LQTIDVDSGL PVYAPLEVTV1440
1441 KETKLYSETK FCEITPCILP ERAILKRICV CGPRYWPQQI ELVFGLRTLG ESNLIANSHR1500
1501 ELDSDSVDHL VSTFSSDPSL IAFAQLCCDK SWNNSFDFLI LDLILWSQVA LAYNEAVSTG1560
1561 RLASSGGFVQ SIFLASLRKR CEEVLNCSTE LKINLRNYLT SEAWPYDKNS KLQKDIIILS1620
1621 WYLKWFNVPS PSIIKAAVEK IKSKSKNSTS AIPLLRLLLP NTHISVIGEI DRVFFPSN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
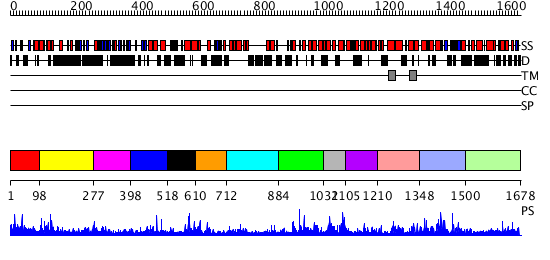
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..97] | 1.013977 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [98..276] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [277..397] | 1.042994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [398..517] | 1.041996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [518..609] | 1.102995 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [610..711] | 2.124989 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [712..883] | 1.124982 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [884..1031] | 1.132986 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1032..1104] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1105..1209] | 2.137995 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1210..1347] | 3.134993 | View MSA. No confident structure predictions are available. | |
| 12 | View Details | [1348..1499] | 3.135985 | View MSA. No confident structure predictions are available. | |
| 13 | View Details | [1500..1678] | 1.062991 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|