













| General Information: |
|
| Name(s) found: |
FTSH2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 695 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
thylakoid lumen [IDA] membrane [IDA] chloroplast envelope [IDA] chloroplast thylakoid membrane [IDA] thylakoid [IDA] |
| Biological Process: |
oxygen and reactive oxygen species metabolic process
[IDA]
protein catabolic process [IDA] thylakoid membrane organization [IMP] photoinhibition [IMP] PSII associated light-harvesting complex II catabolic process [TAS] |
| Molecular Function: |
ATPase activity
[ISS]
zinc ion binding [ISS] ATP-dependent peptidase activity [ISS] metallopeptidase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAASSACLVG NGLSVNTTTK QRLSKHFSGR QTSFSSVIRT SKVNVVKASL DGKKKQEGRR 60
61 DFLKILLGNA GVGLVASGKA NADEQGVSSS RMSYSRFLEY LDKDRVNKVD LYENGTIAIV 120
121 EAVSPELGNR VERVRVQLPG LSQELLQKLR AKNIDFAAHN AQEDQGSVLF NLIGNLAFPA 180
181 LLIGGLFLLS RRSGGGMGGP GGPGNPLQFG QSKAKFQMEP NTGVTFDDVA GVDEAKQDFM 240
241 EVVEFLKKPE RFTAVGAKIP KGVLLIGPPG TGKTLLAKAI AGEAGVPFFS ISGSEFVEMF 300
301 VGVGASRVRD LFKKAKENAP CIVFVDEIDA VGRQRGTGIG GGNDEREQTL NQLLTEMDGF 360
361 EGNTGVIVVA ATNRADILDS ALLRPGRFDR QVSVDVPDVK GRTDILKVHA GNKKFDNDVS 420
421 LEIIAMRTPG FSGADLANLL NEAAILAGRR ARTSISSKEI DDSIDRIVAG MEGTVMTDGK 480
481 SKSLVAYHEV GHAVCGTLTP GHDAVQKVTL IPRGQARGLT WFIPSDDPTL ISKQQLFARI 540
541 VGGLGGRAAE EIIFGDSEVT TGAVGDLQQI TGLARQMVTT FGMSDIGPWS LMDSSAQSDV 600
601 IMRMMARNSM SEKLAEDIDS AVKKLSDSAY EIALSHIKNN REAMDKLVEV LLEKETIGGD 660
661 EFRAILSEFT EIPPENRVPS STTTTPASAP TPAAV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
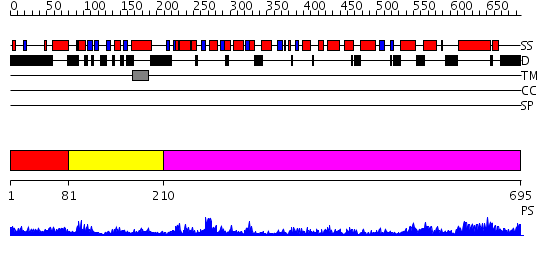
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [81..209] | 17.207608 | No description for PF06480.6 was found. No confident structure predictions are available. | |
| 3 | View Details | [210..695] | 126.0 | EDTA treated |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.40 |
Source: Reynolds et al. (2008)