













| General Information: |
|
| Name(s) found: |
TC132_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1206 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast outer membrane
[IDA][ISS]
membrane [IDA] |
| Biological Process: |
protein targeting to chloroplast
[IGI]
|
| Molecular Function: |
transmembrane receptor activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGDGTEFVVR SDREDKKLAE DRISDEQVVK NELVRSDEVR DDNEDEVFEE AIGSENDEQE 60
61 EEEDPKRELF ESDDLPLVET LKSSMVEHEV EDFEEAVGDL DETSSNEGGV KDFTAVGESH 120
121 GAGEAEFDVL ATKMNGDKGE GGGGGSYDKV ESSLDVVDTT ENATSTNTNG SNLAAEHVGI 180
181 ENGKTHSFLG NGIASPKNKE VVAEVIPKDD GIEEPWNDGI EVDNWEERVD GIQTEQEVEE 240
241 GEGTTENQFE KRTEEEVVEG EGTSKNLFEK QTEQDVVEGE GTSKDLFENG SVCMDSESEA 300
301 ERNGETGAAY TSNIVTNASG DNEVSSAVTS SPLEESSSGE KGETEGDSTC LKPEQHLASS 360
361 PHSYPESTEV HSNSGSPGVT SREHKPVQSA NGGHDVQSPQ PNKELEKQQS SRVHVDPEIT 420
421 ENSHVETEPE VVSSVSPTES RSNPAALPPA RPAGLGRASP LLEPASRAPQ QSRVNGNGSH 480
481 NQFQQAEDST TTEADEHDET REKLQLIRVK FLRLAHRLGQ TPHNVVVAQV LYRLGLAEQL 540
541 RGRNGSRVGA FSFDRASAMA EQLEAAGQDP LDFSCTIMVL GKSGVGKSAT INSIFDEVKF 600
601 CTDAFQMGTK RVQDVEGLVQ GIKVRVIDTP GLLPSWSDQA KNEKILNSVK AFIKKNPPDI 660
661 VLYLDRLDMQ SRDSGDMPLL RTISDVFGPS IWFNAIVGLT HAASVPPDGP NGTASSYDMF 720
721 VTQRSHVIQQ AIRQAAGDMR LMNPVSLVEN HSACRTNRAG QRVLPNGQVW KPHLLLLSFA 780
781 SKILAEANAL LKLQDNIPGR PFAARSKAPP LPFLLSSLLQ SRPQPKLPEQ QYGDEEDEDD 840
841 LEESSDSDEE SEYDQLPPFK SLTKAQMATL SKSQKKQYLD EMEYREKLLM KKQMKEERKR 900
901 RKMFKKFAAE IKDLPDGYSE NVEEESGGPA SVPVPMPDLS LPASFDSDNP THRYRYLDSS 960
961 NQWLVRPVLE THGWDHDIGY EGVNAERLFV VKEKIPISVS GQVTKDKKDA NVQLEMASSV1020
1021 KHGEGKSTSL GFDMQTVGKE LAYTLRSETR FNNFRRNKAA AGLSVTHLGD SVSAGLKVED1080
1081 KFIASKWFRI VMSGGAMTSR GDFAYGGTLE AQLRDKDYPL GRFLTTLGLS VMDWHGDLAI1140
1141 GGNIQSQVPI GRSSNLIARA NLNNRGAGQV SVRVNSSEQL QLAMVAIVPL FKKLLSYYYP1200
1201 QTQYGQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
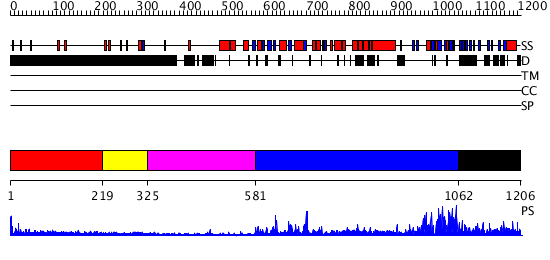
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..218] | 2.321996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [219..324] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [325..580] | 28.69897 | Structure of the GTP-binding protein TrmE from Thermotoga maritima | |
| 4 | View Details | [581..1061] | 69.0 | No description for 2hjgA was found. | |
| 5 | View Details | [1062..1206] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)