













| General Information: |
|
| Name(s) found: |
BGAL2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 727 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apoplast
[IDA]
|
| Biological Process: |
carbohydrate metabolic process
[IEA]
|
| Molecular Function: |
catalytic activity
[IEA]
cation binding [IEA] beta-galactosidase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSMHFRNKAW IILAILCFSS LIHSTEAVVT YDHKALIING QRRILISGSI HYPRSTPEMW 60
61 PDLIKKAKEG GLDVIQTYVF WNGHEPSPGN YYFQDRYDLV KFTKLVHQAG LYLDLRIGPY 120
121 VCAEWNFGGF PVWLKYVPGM VFRTDNEPFK IAMQKFTKKI VDMMKEEKLF ETQGGPIILS 180
181 QIENEYGPMQ WEMGAAGKAY SKWTAEMALG LSTGVPWIMS KQEDAPYPII DTCNGFYCEG 240
241 FKPNSDNKPK LWTENWTGWF TEFGGAIPNR PVEDIAFSVA RFIQNGGSFM NYYMYYGGTN 300
301 FDRTAGVFIA TSYDYDAPID EYGLLREPKY SHLKELHKVI KLCEPALVSV DPTITSLGDK 360
361 QEIHVFKSKT SCAAFLSNYD TSSAARVMFR GFPYDLPPWS VSILPDCKTE YYNTAKIRAP 420
421 TILMKMIPTS TKFSWESYNE GSPSSNEAGT FVKDGLVEQI SMTRDKTDYF WYFTDITIGS 480
481 DESFLKTGDN PLLTIFSAGH ALHVFVNGLL AGTSYGALSN SKLTFSQNIK LSVGINKLAL 540
541 LSTAVGLPNA GVHYETWNTG ILGPVTLKGV NSGTWDMSKW KWSYKIGLRG EAMSLHTLAG 600
601 SSAVKWWIKG FVVKKQPLTW YKSSFDTPRG NEPLALDMNT MGKGQVWVNG HNIGRHWPAY 660
661 TARGNCGRCN YAGIYNEKKC LSHCGEPSQR WYHVPRSWLK PFGNLLVIFE EWGGDPSGIS 720
721 LVKRTAK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
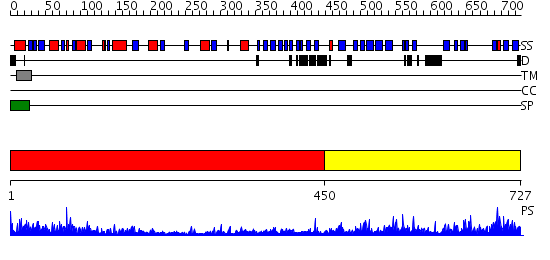
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..449] | 50.69897 | Native structure of beta-galactosidase from Penicillium sp. | |
| 2 | View Details | [450..727] | 2.14 | beta-Glucuronidase; beta-Glucuronidase, domain 3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|