













| General Information: |
|
| Name(s) found: |
UPL5_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 873 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
|
| Biological Process: |
protein modification process
[ISS]
senescence [IMP] |
| Molecular Function: |
ubiquitin-protein ligase activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTLSRSSADD STNNANRSYS AVAGTDNKRK RDEDSSDYVG VAESLEMLKK QEIDADHMAA 60
61 SAQQTLISWR SGENSRSLSS SGECSSSNRP ESTRLQIFVR MMSGGKTIVI HAEKYDTVEK 120
121 LHQRIEWKTK IPALEQRVIY KGKQLQRENS LTYYSIEQDA SLQLVARMQS TEHPVAWQTI 180
181 DDIMYTISRM YKGENLQSNI NEKIVTFFAM IPVESDESIA KYLNIFSNSS VPAALVMLYA 240
241 SSLERNKSCA KSSVKLFLSN CVALPKNQKN YCLPIVLEFC KLLRKVCPDQ KLYVTCRNTL 300
301 GSMLETFDNP HGVYNDQYET FGVEIFPFFT ELTGLLLNEL AQNSGPSFCD FQKVSSFWQQ 360
361 LRKVIELKVA FPIPIVLPMQ STALEAEIRH LHRLFGSLLT TMDLCMCRVE SSLADKEVGN 420
421 SETMSSSWSQ YLSILKIINS MSNIYQGAKG QLAVMLNKNK VSFSALVVKF AKRGDDHQWI 480
481 FEYKEATNFE ARRHLAMLLF PDVKEDFEEM HEMLIDRSNL LSESFEYIVG ASPEALHGGL 540
541 FMEFKNEEAT GPGVLREWFY LVCQEIFNPK NTLFLRSADD FRRFSPNPAS KVDPLHPDFF 600
601 EFTGRVIALA LMHKVQVGVL FDRVFFLQLA GLKISLEDIK DTDRIMYNSC KQILEMDPEF 660
661 FDSNAGLGLT FVLETEELGK RDTIELCPDG KLKAVNSKNR KQYVDLLIER RFATPILEQV 720
721 KQFSRGFTDM LSHSVPPRSF FKRLYLEDLD GMLRGGENPI SIDDWKAHTE YNGFKETDRQ 780
781 IDWFWKILKK MTEEEQRSIL FFWTSNKFVP VEGFRGLSSK LYIYRLYEAN DRLPLSHTCF 840
841 YRLCIPRYPT ITLMEQRLRL IAQDHVSSSF GKW |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
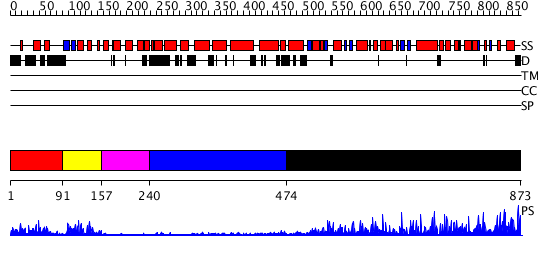
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | 20.09691 | No description for 2ojrA was found. | |
| 2 | View Details | [91..156] | 18.09691 | X-ray Crystal Structure of a Chemically Synthesized [D-Gln35]Ubiquitin | |
| 3 | View Details | [157..239] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [240..473] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [474..873] | 116.0 | Regulation of Smurf2 Ubiquitin Ligase Activity by Anchoring the E2 to the HECT domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)