













| General Information: |
|
| Name(s) found: |
APRR9_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 468 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to temperature stimulus
[IGI]
red or far-red light signaling pathway [IMP] circadian rhythm [IMP][TAS] |
| Molecular Function: |
DNA binding
[IPI]
protein binding [IPI] transcription regulator activity [TAS] transcription repressor activity [IDA] two-component response regulator activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGEIVVLSSD DGMETIKNRV KSSEVVQWEK YLPKTVLRVL LVESDYSTRQ IITALLRKCC 60
61 YKVVAVSDGL AAWEVLKEKS HNIDLILTEL DLPSISGFAL LALVMEHEAC KNIPVIMMSS 120
121 QDSIKMVLKC MLRGAADYLI KPMRKNELKN LWQHVWRRLT LRDDPTAHAQ SLPASQHNLE 180
181 DTDETCEDSR YHSDQGSGAQ AINYNGHNKL MENGKSVDER DEFKETFDVT MDLIGGIDKR 240
241 PDSIYKDKSR DECVGPELGL SLKRSCSVSF ENQDESKHQK LSLSDASAFS RFEESKSAEK 300
301 AVVALEESTS GEPKTPTESH EKLRKVTSDQ GSATTSSNQE NIGSSSVSFR NQVLQSTVTN 360
361 QKQDSPIPVE SNREKAASKE VEAGSQSTNE GIAGQSSSTE KPKEEESAKQ RWSRSQREAA 420
421 LMKFRLKRKD RCFDKKVRYQ SRKKLAEQRP RVKGQFVRTV NSDASTKS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
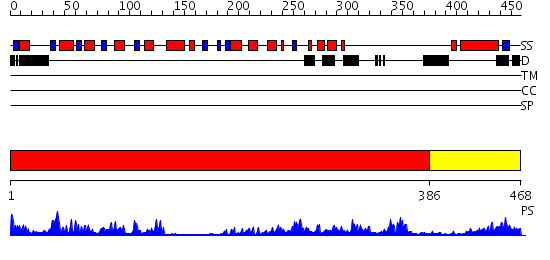
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..385] | 52.30103 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 2 | View Details | [386..468] | 40.39794 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)