













| General Information: |
|
| Name(s) found: |
SM3L2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1029 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
DNA mediated transformation
[IMP]
|
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA][ISS] DNA binding [IEA][ISS] zinc ion binding [IEA][ISS] protein binding [IEA][ISS] hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides [IEA] ATP-dependent helicase activity [IEA] helicase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTKVSDDLV STVRSVVGSD YSDMDIIRAL HMANHDPTAA INIIFDTPSF AKPDVATPTP 60
61 SGSNGGKRVD SGLKGCTFGD SGSVGANHRV EEENESVNGG GEESVSGNEW WFVGCSELAG 120
121 LSTCKGRKLK SGDELVFTFP HSKGLKPETT PGKRGFGRGR PALRGASDIV RFSTKDSGEI 180
181 GRIPNEWARC LLPLVRDKKI RIEGSCKSAP EALSIMDTIL LSVSVYINSS MFQKHSATSF 240
241 KTASNTAEES MFHPLPNLFR LLGLIPFKKA EFTPEDFYSK KRPLSSKDGS AIPTSLLQLN 300
301 KVKNMNQDAN GDENEQCISD GDLDNIVGVG DSSGLKEMET PHTLLCELRP YQKQALHWMT 360
361 QLEKGNCTDE AATMLHPCWE AYCLADKREL VVYLNSFTGD ATIHFPSTLQ MARGGILADA 420
421 MGLGKTVMTI SLLLAHSWKA ASTGFLCPNY EGDKVISSSV DDLTSPPVKA TKFLGFDKRL 480
481 LEQKSVLQNG GNLIVCPMTL LGQWKTEIEM HAKPGSLSVY VHYGQSRPKD AKLLSQSDVV 540
541 ITTYGVLTSE FSQENSADHE GIYAVRWFRI VLDEAHTIKN SKSQISLAAA ALVADRRWCL 600
601 TGTPIQNNLE DLYSLLRFLR IEPWGTWAWW NKLVQKPFEE GDERGLKLVQ SILKPIMLRR 660
661 TKSSTDREGR PILVLPPADA RVIYCELSES ERDFYDALFK RSKVKFDQFV EQGKVLHNYA 720
721 SILELLLRLR QCCDHPFLVM SRGDTAEYSD LNKLSKRFLS GKSSGLEREG KDVPSEAFVQ 780
781 EVVEELRKGE QGECPICLEA LEDAVLTPCA HRLCRECLLA SWRNSTSGLC PVCRNTVSKQ 840
841 ELITAPTESR FQVDVEKNWV ESSKITALLE ELEGLRSSGS KSILFSQWTA FLDLLQIPLS 900
901 RNNFSFVRLD GTLSQQQREK VLKEFSEDGS ILVLLMSLKA GGVGINLTAA SNAFVMDPWW 960
961 NPAVEEQAVM RIHRIGQTKE VKIRRFIVKG TVEERMEAVQ ARKQRMISGA LTDQEVRSAR1020
1021 IEELKMLFT |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
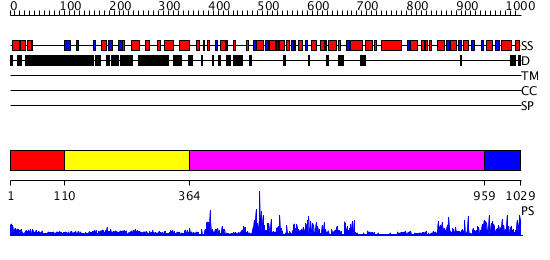
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [110..363] | 10.30103 | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and dimethylarginine 2 | |
| 3 | View Details | [364..958] | 71.221849 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | |
| 4 | View Details | [959..1029] | 31.0 | Sulfolobus solfataricus SWI2/SNF2 ATPase C-terminal domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)