













| General Information: |
|
| Name(s) found: |
ANXD2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 317 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytosol [IDA] thylakoid [IDA] cell surface [NAS] |
| Biological Process: |
polysaccharide transport
[TAS]
response to cold [IEP] response to heat [IEP] response to salt stress [IEP] response to water deprivation [IEP] |
| Molecular Function: |
calcium ion binding
[IEA][ISS]
calcium-dependent phospholipid binding [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASLKVPSNV PLPEDDAEQL HKAFSGWGTN EKLIISILAH RNAAQRSLIR SVYAATYNED 60
61 LLKALDKELS SDFERAVMLW TLDPPERDAY LAKESTKMFT KNNWVLVEIA CTRPALELIK 120
121 VKQAYQARYK KSIEEDVAQH TSGDLRKLLL PLVSTFRYEG DDVNMMLARS EAKILHEKVS 180
181 EKSYSDDDFI RILTTRSKAQ LGATLNHYNN EYGNAINKNL KEESDDNDYM KLLRAVITCL 240
241 TYPEKHFEKV LRLSINKMGT DEWGLTRVVT TRTEVDMERI KEEYQRRNSI PLDRAIAKDT 300
301 SGDYEDMLVA LLGHGDA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
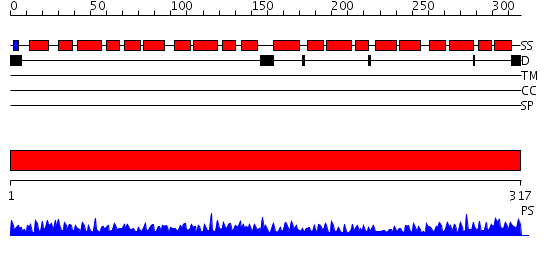
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..317] | 100.0 | X-RAY STRUCTURE OF ANNEXIN FROM ARABIDOPSIS THALIANA GENE AT1G35720 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)