













| General Information: |
|
| Name(s) found: |
gi|25406340
[NCBI NR]
gi|15221221 [NCBI NR] gi|12324793 [NCBI NR] |
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 753 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
RNA splicing
[IEA][ISS]
RNA-dependent DNA replication [IEA][ISS] |
| Molecular Function: |
RNA-directed DNA polymerase activity
[IEA][ISS]
RNA binding [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASKETGMFS LAGELASLVE ESSSHVDDDS KPRSRMELKR SLELRLKKRV KEQCINGKFS 60
61 DLLKKVIARP ETLRDAYDCI RLNSNVSITE RNGSVAFDSI AEELSSGVFD VASNTFSIVA 120
121 RDKTKEVLVL PSVALKVVQE AIRIVLEVVF SPHFSKISHS CRSGRGRASA LKYINNNISR 180
181 SDWCFTLSLN KKLDVSVFEN LLSVMEEKVE DSSLSILLRS MFEARVLNLE FGGFPKGHGL 240
241 PQEGVLSRVL MNIYLDRFDH EFYRISMRHE ALGLDSKTDE DSPGSKLRSW FRRQAGEQGL 300
301 KSTTEQDVAL RVYCCRFMDE IYFSVSGPKK VASDIRSEAI GFLRNSLHLD ITDETDPSPC 360
361 EATSGLRVLG TLVRKNVRES PTVKAVHKLK EKVRLFALQK EEAWTLGTVR IGKKWLGHGL 420
421 KKVKESEIKG LADSNSTLSQ ISCHRKAGME TDHWYKILLR IWMEDVLRTS ADRSEEFVLS 480
481 KHVVEPTVPQ ELRDAFYKFQ NAAAAYVSSE TANLEALLPC PQSHDRPVFF GDVVAPTNAI 540
541 GRRLYRYGLI TAKGYARSNS MLILLDTAQI IDWYSGLVRR WVIWYEGCSN FDEIKALIDN 600
601 QIRMSCIRTL AAKYRIHENE IEKRLDLELS TIPSAEDIEQ EIQHEKLDSP AFDRDEHLTY 660
661 GLSNSGLCLL SLARLVSESR PCNCFVIGCS MAAPAVYTLH AMERQKFPGW KTGFSVCIPS 720
721 SLNGRRIGLC KQHLKDLYIG QISLQAVDFG AWR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
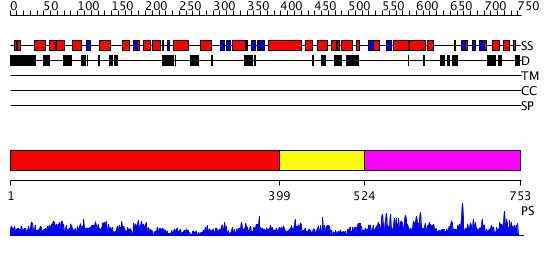
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..398] | 2.14 | Crystal structure of RNA-dependent RNA Polymerase 3D from human rhinovirus serotype 16 | |
| 2 | View Details | [399..523] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [524..753] | 5.958607 | No description for PF01348.12 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)