













| General Information: |
|
| Name(s) found: |
SERB2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 422 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
response to cadmium ion
[IEP]
|
| Molecular Function: |
pyridoxal phosphate binding
[IEA]
catalytic activity [IEA] O-phospho-L-serine:2-oxoglutarate aminotransferase activity [ISS] transaminase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAASTNSFLI GNQTQIPSLK PKSISQSFIH FTKPNTINLT TRTKSVSIRC ASASTTVGSE 60
61 QRVINFAAGP AALPENVLLK AQSDLYNWRG SGMSVMEMSH RGKEFLSIIQ KAESDLRQLL 120
121 EIPSEYSVLF LQGGATTQFA ALPLNLCKSD DSVDYIVTGS WGDKAFKEAK KYCNPKVIWS 180
181 GKSEKYTKVP TFDGLEQSSD AKYLHICANE TIHGVEFKDY PLVENPDGVL IADMSSNFCS 240
241 KPVDVSKFGV IYAGAQKNVG PSGVTIVIIR KDLIGNARDI TPVMLDYKIH DENSSLYNTP 300
301 PCFGIYMCGL VFDDLLEQGG LKEVEKKNQR KAELLYNAID ESRGFFRCPV EKSVRSLMNV 360
361 PFTLEKSELE AEFIKEAAKE KMVQLKGHRS VGGMRASIYN AMPLAGVEKL VAFMKDFQAR 420
421 HA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
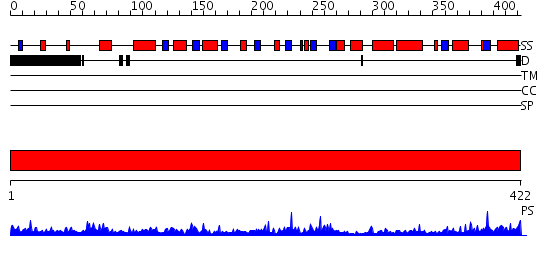
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..422] | 66.39794 | Phosphoserine aminotransferase, PSAT |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)