













| General Information: |
|
| Name(s) found: |
SM3L3_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1277 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast envelope
[IDA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA]
DNA binding [IEA][ISS] ATP binding [IEA][ISS] zinc ion binding [IEA][ISS] protein binding [IEA][ISS] hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides [IEA] ATP-dependent helicase activity [IEA] helicase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAIVDDAEMR LTESEAVSSS DDRKIVADTP DFIDESSLVI RTTTGVRISA LPAEQSLVDS 60
61 DGSNSEVTLP AKDEVISDGF TCVNKEIVES DSFREQNLEI GEPDLDVENR KEAMIIDSIE 120
121 NSVVEIVSSA SGDDCNVKVE VVEPELLVEN LVVAKEEEEM IVDSIEDSVV EIVSTASGCD 180
181 CNVKVEVVDP ELCVDNLVVV KEEEMIADSI AESVVETVSR GLDYECVDVK VKEEPDLGTK 240
241 LEEDSVFPNV LEKKDEVIKV LEDQPSEINK KLEQENDDLF SSGDSDGTSA KRRKMEMESY 300
301 APVGVESCIL APTPLRVVKP EKLDTPEVID LESEKSYTHV KMEPVEEIKV EAVKMSSQVE 360
361 DVKFSREQKS VYVKKEPVGA RKVKVEDGDF PVEKDWYLVG RSLVTATSTS KGRKLEDNEI 420
421 VNFTFSSVAK WKVPNIVRFS TKRCGEIGRL PMEWSNWAVS LLRSGKVKML GRCVAAPPFL 480
481 TMMQEIMLYV SFYIHSSIFT DVSKSTWRIG SSPNLESTLH PLLQLFKHLT IKPYQKAEFT 540
541 PEELNSRKRS LNLEDDYDER AALLAIAKRR KGCQQSLEQN KDEEEAPESY MNRVVGAADS 600
601 YNLEEMEAPS TLTCNLRPYQ KQALYWMSES EKGIDVEKAA ETLHPCWEAY RICDERAPSI 660
661 YLNIFSGEAT IQFPTATQMA RGGILADAMG LGKTVMTIAL ILARPGRGNP ENEDVLVADV 720
721 NADKRNRKEI HMALTTVKAK GGTLIICPMA LLSQWKDELE THSKPDTVSV LVYYGGDRTH 780
781 DAKAIASHDV VLTTYGVLTS AYKQDMANSI FHRIDWYRIV LDEAHTIKSW KTQAAKATFE 840
841 LSSHCRWCLT GTPLQNKLED LYSLLCFLHV EPWCNWAWWS KLIQKPYENG DPRGLKLIKA 900
901 ILRPLMLRRT KETRDKEGSL ILELPPTDVQ VIECEQSEAE RDFYTALFKR SKVQFDQFVA 960
961 QGKVLHNYAN ILELLLRLRQ CCNHPFLVMS RADSQQYADL DSLARRFLDN NPDSVSQNAP1020
1021 SRAYIEEVIQ DLRDGNSKEC PICLESADDP VLTPCAHRMC RECLLTSWRS PSCGLCPICR1080
1081 TILKRTELIS CPTDSIFRVD VVKNWKESSK VSELLKCLEK IKKSGSGEKS IVFSQWTSFL1140
1141 DLLEIPLRRR GFEFLRFDGK LAQKGREKVL KEFNETKQKT ILLMSLKAGG VGLNLTAASS1200
1201 VFLMDPWWNP AVEEQAIMRI HRIGQKRTVF VRRFIVKDTV EERMQQVQAR KQRMIAGALT1260
1261 DEEVRSARLE ELKMLFR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
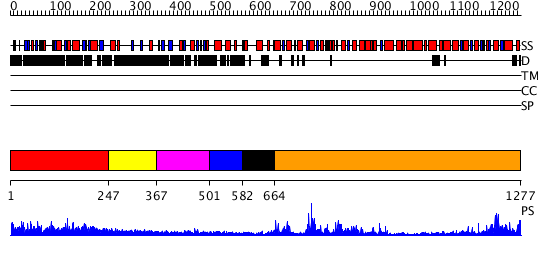
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..246] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [247..366] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [367..500] | 19.769551 | No description for PF08797.2 was found. No confident structure predictions are available. | |
| 4 | View Details | [501..581] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [582..663] | 66.69897 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | |
| 6 | View Details | [664..1277] | 48.39794 | No description for 2fdcA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)