













| General Information: |
|
| Name(s) found: |
RFS5_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 783 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
sucrose biosynthetic process
[IMP]
mannitol biosynthetic process [IMP] raffinose family oligosaccharide biosynthetic process [IMP] response to water deprivation [IEP] |
| Molecular Function: |
galactinol-sucrose galactosyltransferase activity
[ISS]
hydrolase activity, hydrolyzing O-glycosyl compounds [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASPCLTKSD SGINGVDFTE KFRLEDSTLL ANGQVVLTDV PVNVTLTSSP YLVDKDGVPL 60
61 DVSAGSFIGF NLDGEPKSHH VASIGKLKNI RFMSIFRFKV WWTTHWVGSN GRDIENETQI 120
121 IILDQSGSDS GPGSGSGRPY VLLLPLLEGS FRSSFQSGED DDVAVCVESG STEVTGSEFR 180
181 QIVYVHAGDD PFKLVKDAMK VIRVHMNTFK LLEEKSPPGI VDKFGWCTWD AFYLTVNPDG 240
241 VHKGVKCLVD GGCPPGLVLI DDGWQSIGHD SDGIDVEGMN ITVAGEQMPC RLLKFEENHK 300
301 FKDYVSPKDQ NDVGMKAFVR DLKDEFSTVD YIYVWHALCG YWGGLRPEAP ALPPSTIIRP 360
361 ELSPGLKLTM EDLAVDKIIE TGIGFASPDL AKEFYEGLHS HLQNAGIDGV KVDVIHILEM 420
421 LCQKYGGRVD LAKAYFKALT SSVNKHFNGN GVIASMEHCN DFMFLGTEAI SLGRVGDDFW 480
481 CTDPSGDPNG TFWLQGCHMV HCAYNSLWMG NFIQPDWDMF QSTHPCAEFH AASRAISGGP 540
541 IYISDCVGKH DFDLLKRLVL PNGSILRCEY YALPTRDRLF EDPLHDGKTM LKIWNLNKYT 600
601 GVIGAFNCQG GGWCRETRRN QCFSECVNTL TATTSPKDVE WNSGSSPISI ANVEEFALFL 660
661 SQSKKLLLSG LNDDLELTLE PFKFELITVS PVVTIEGNSV RFAPIGLVNM LNTSGAIRSL 720
721 VYNDESVEVG VFGAGEFRVY ASKKPVSCLI DGEVVEFGYE DSMVMVQVPW SGPDGLSSIQ 780
781 YLF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
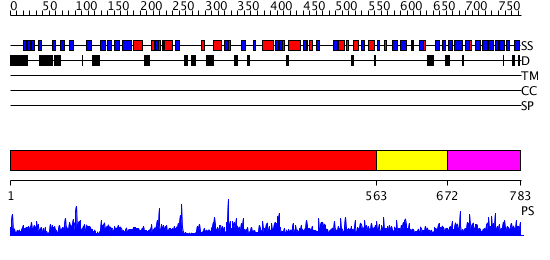
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..562] | 71.09691 | Crystal structure of Alpha-galactosidase (EC 3.2.1.22) (Melibiase) (tm1192) from Thermotoga maritima at 2.34 A resolution | |
| 2 | View Details | [563..671] | 10.30103 | Melibiase | |
| 3 | View Details | [672..783] | 2.041982 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)