













| General Information: |
|
| Name(s) found: |
IPYR4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 216 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
membrane [ISS] |
| Biological Process: |
metabolic process
[ISS]
response to cadmium ion [IEP] |
| Molecular Function: |
inorganic diphosphatase activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPPIEVSTK SYVEKHVSLP TLNERILSSM SHRSVAAHPW HDLEIGPEAP IIFNCVVEIG 60
61 KGSKVKYELD KTTGLIKVDR ILYSSVVYPH NYGFIPRTLC EDSDPIDVLV IMQEPVIPGC 120
121 FLRAKAIGLM PMIDQGEKDD KIIAVCADDP EYRHYNDISE LPPHRMAEIR RFFEDYKKNE 180
181 NKEVAVNDFL PATAAYDAVQ HSMDLYADYV VENLRR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
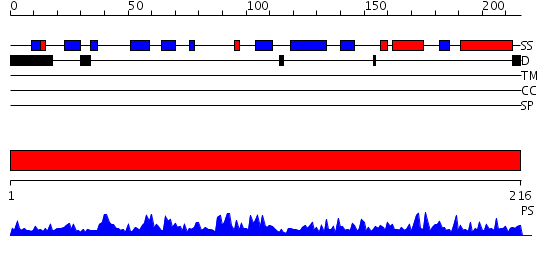
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..216] | 66.522879 | Crystal structure of the Inorganic pyrophosphatase from the hyperthermophilic archaeon Pyrococcus horikoshii OT3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)